Chapter 7
Microbial Genetics
By Boundless

Bacterial genomes are smaller in size (size range from 139 kbp to 13,000 kpb) between species when compared with genomes of eukaryotes.
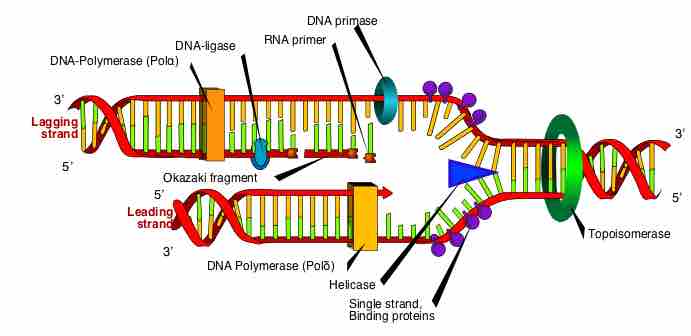
Prokaryotic DNA is replicated by DNA polymerase III in the 5' to 3' direction at a rate of 1000 nucleotides per second.

Gene Inversion utilizes recombinases to invert DNA sequences, resulting in an ON to OFF switch in the gene located within this switch.
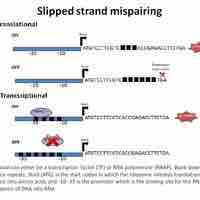
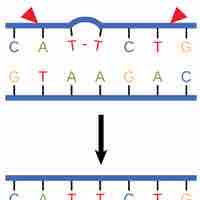
Slipped strand mispairing (SSM) is a process that produces mispairing of short repeat sequences during DNA synthesis.

The nucleoid is an irregularly-shaped region within the cell of a prokaryote that contains all or most of the genetic material.
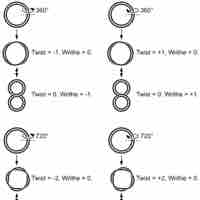
DNA supercoiling refers to the over- or under-winding of a DNA strand, and is an expression of the strain on that strand.

An open reading frame (ORF) is the part of a reading frame that varies in size and content in bacterial genomes.
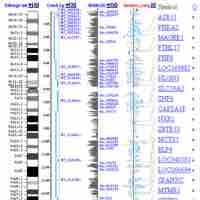
Bioinformatics is the study of methods for storing, retrieving and analyzing biological data.
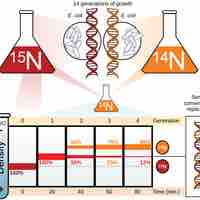
DNA replication uses a semi-conservative method that results in a double-stranded DNA with one parental strand and a new daughter strand.
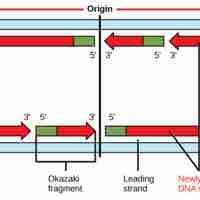
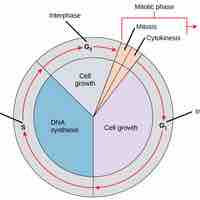
DNA replication in eukaryotes occurs in three stages: initiation, elongation, and termination, which are aided by several enzymes.
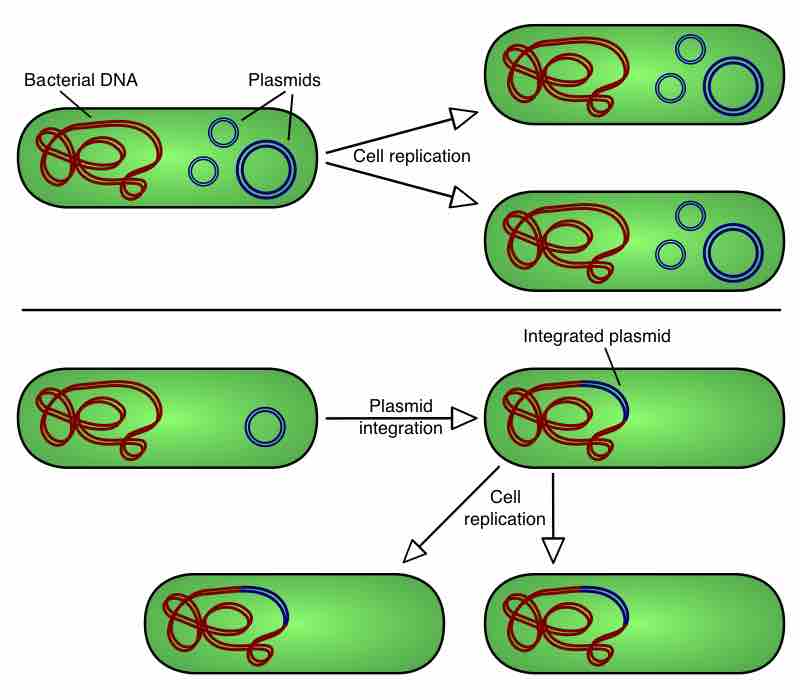
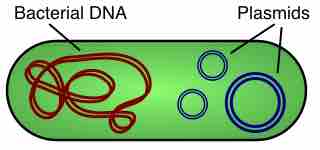
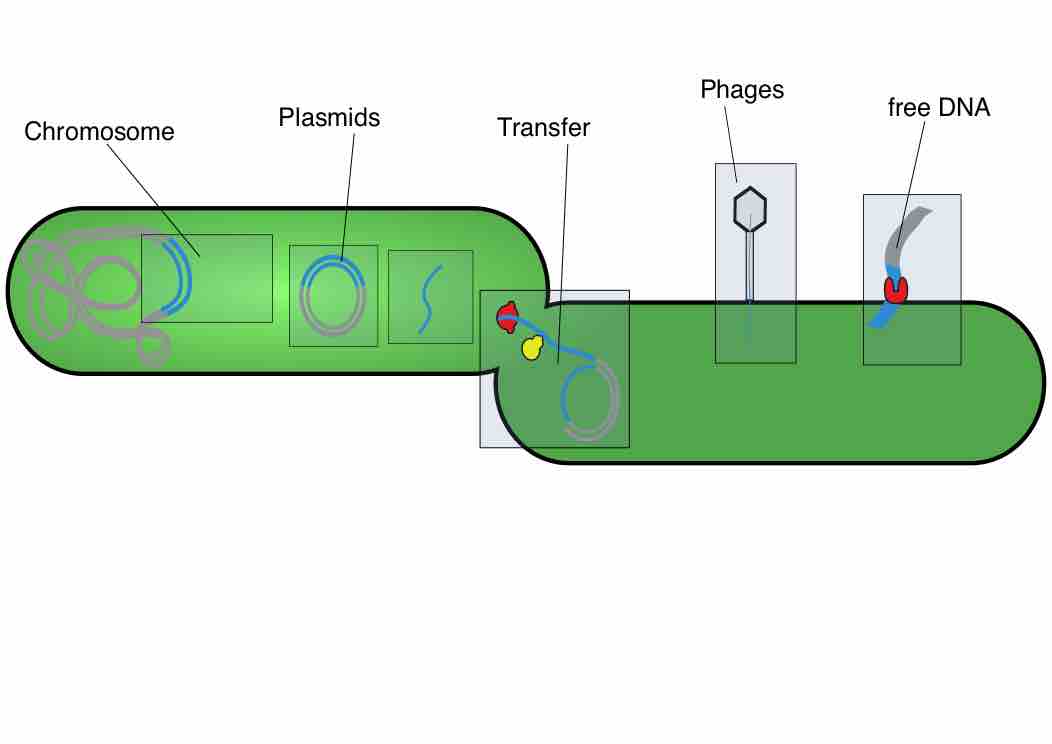
A plasmid is a small DNA molecule that is physically separate from, and can replicate independently of, chromosomal DNA within a cell.
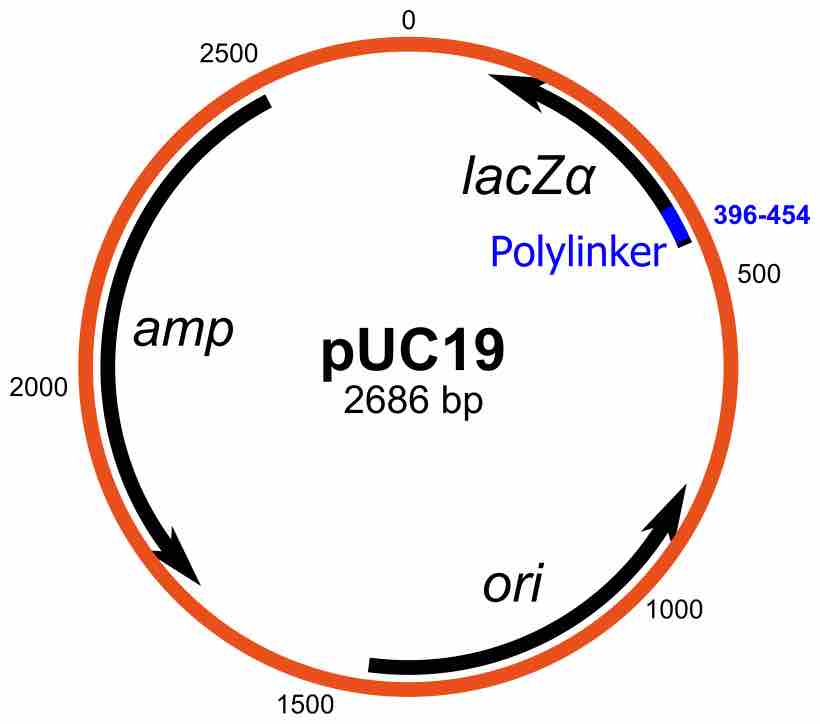
Plasmids are commonly used to multiply (make many copies of) or express particular genes.
Elongation synthesizes pre-mRNA in a 5' to 3' direction, and termination occurs in response to termination sequences and signals.
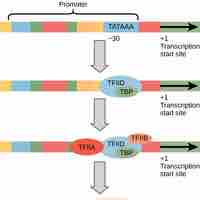
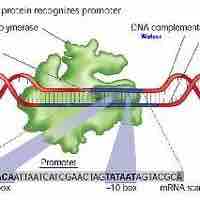
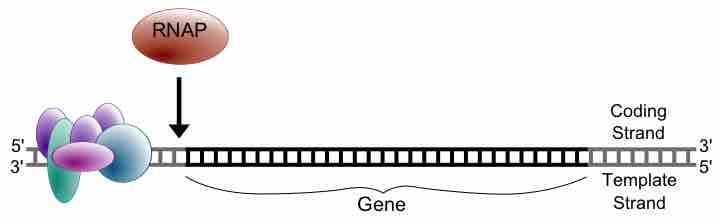
When transcription factors bind to the promoter region, RNA polymerase is placed in an orientation that allows transcription to begin.
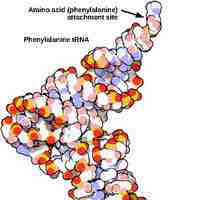
rRNA and tRNA are structural molecules that aid in protein synthesis but are not themselves translated into protein.
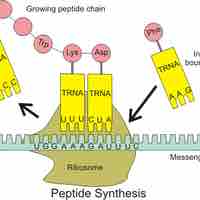
Protein synthesis, or translation of mRNA into protein, occurs with the help of ribosomes, tRNAs, and aminoacyl tRNA synthetases.
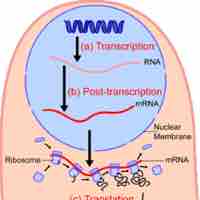
Prokaryotic transcription occurs in the cytoplasm alongside translation and can occur simultaneously.
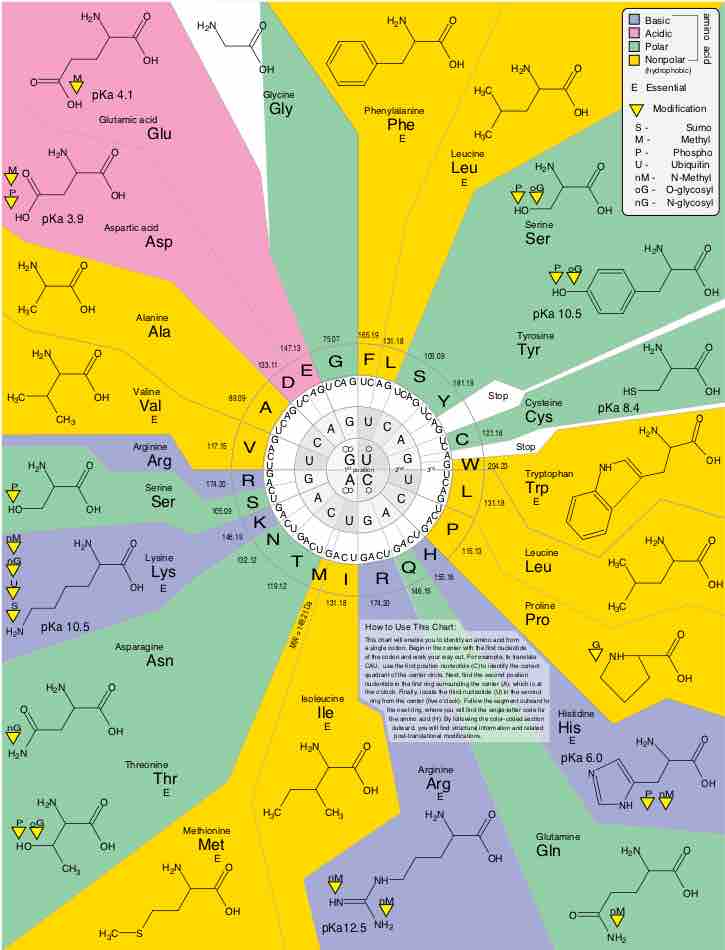
Aside from the 22 standard amino acids, there are many other amino acids that are called non-proteinogenic or non-standard.
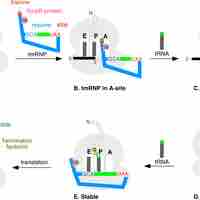
Ribosomes can get stuck on mRNAs, cells have ways of unsticking them.
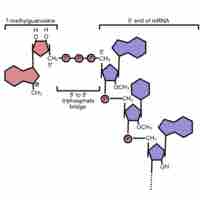
Eukaryotic pre-mRNA receives a 5' cap and a 3' poly (A) tail before introns are removed and the mRNA is considered ready for translation.

Denaturation is a process in which proteins lose their shape and, therefore, their function because of changes in pH or temperature.
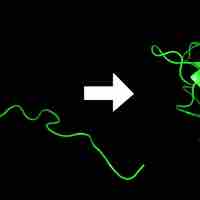
In order to function, proteins must fold into the correct three-dimensional shape, and be targeted to the correct part of the cell.
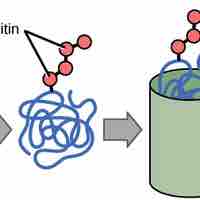
A cell can rapidly change the levels of proteins in response to the environment by adding specific chemical groups to alter gene regulation.
Archaea usually have a single circular chromosome.
Most of the metabolic pathways, which comprise the majority of an organism's genes, are common between Archaea and Bacteria.
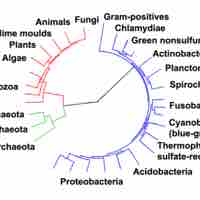
Archaea possess genes and several metabolic pathways that are more closely related to those of eukaryotes than prokaryotes.
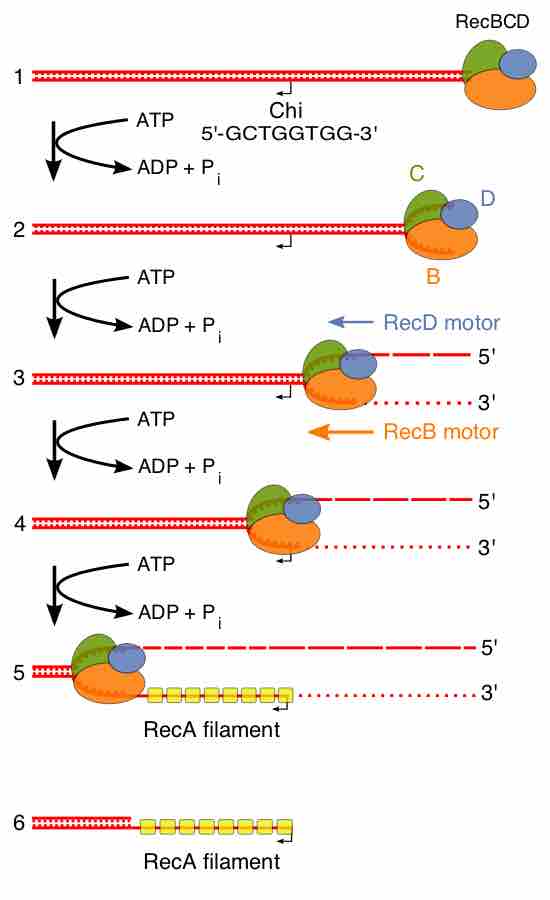
In homologous recombination, a type of genetic recombination, nucleotide sequences are exchanged between two similar molecules of DNA.
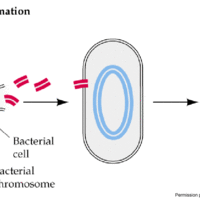
Transformation is the direct uptake, incorporation and expression of exogenous genetic material from its surroundings.
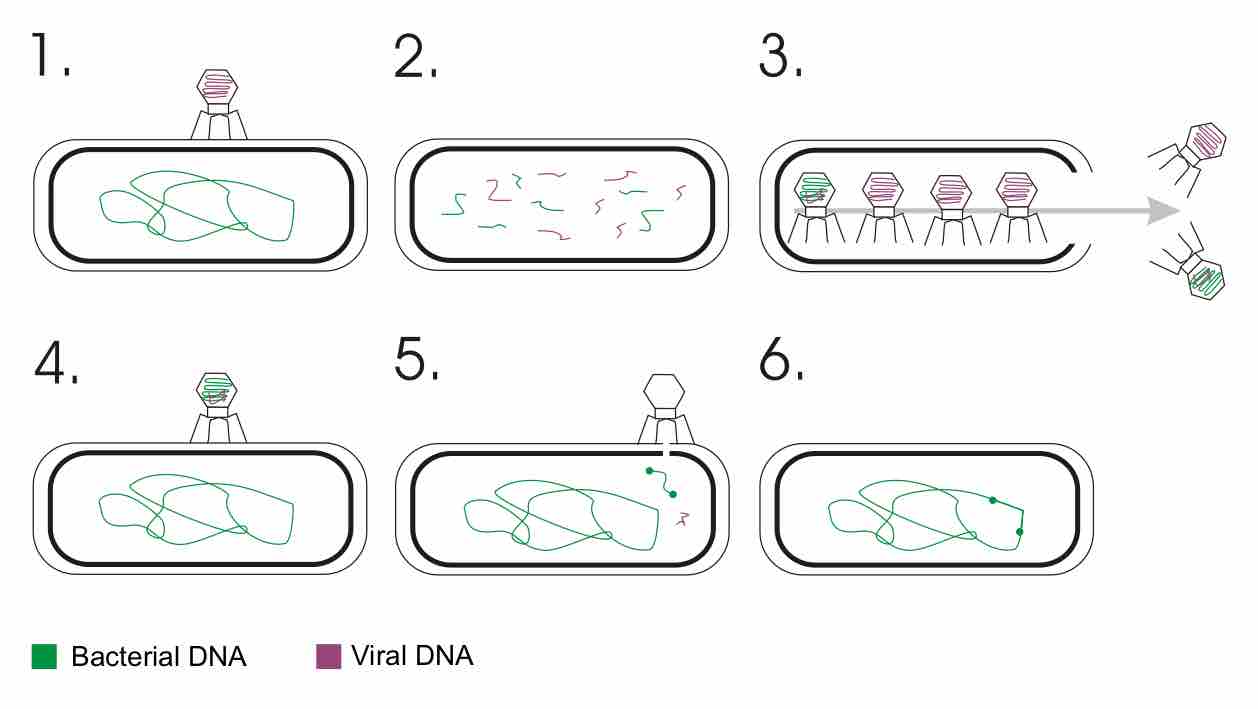
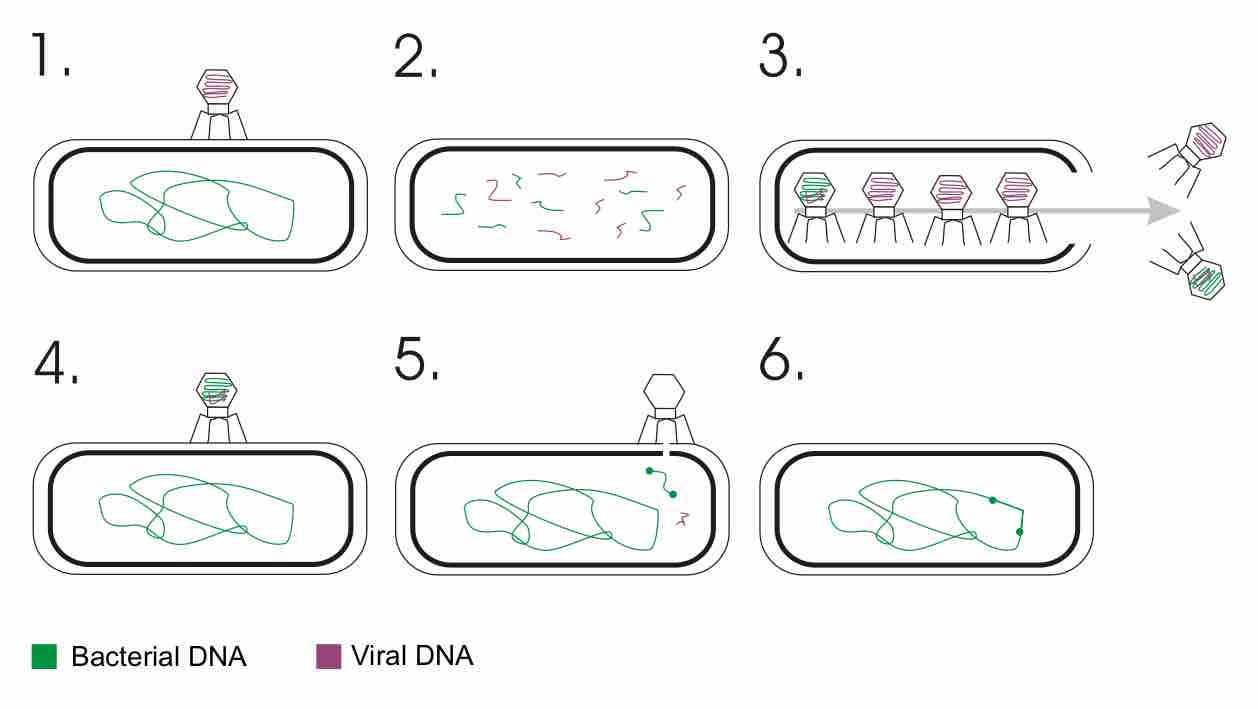
Transduction is the process by which DNA is transferred from one bacterium to another by a virus.
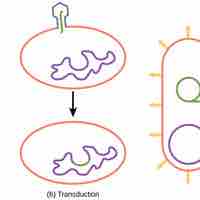
Prokaryotes reproduce asexually by binary fission; they can also exchange genetic material by transformation, transduction, and conjugation.
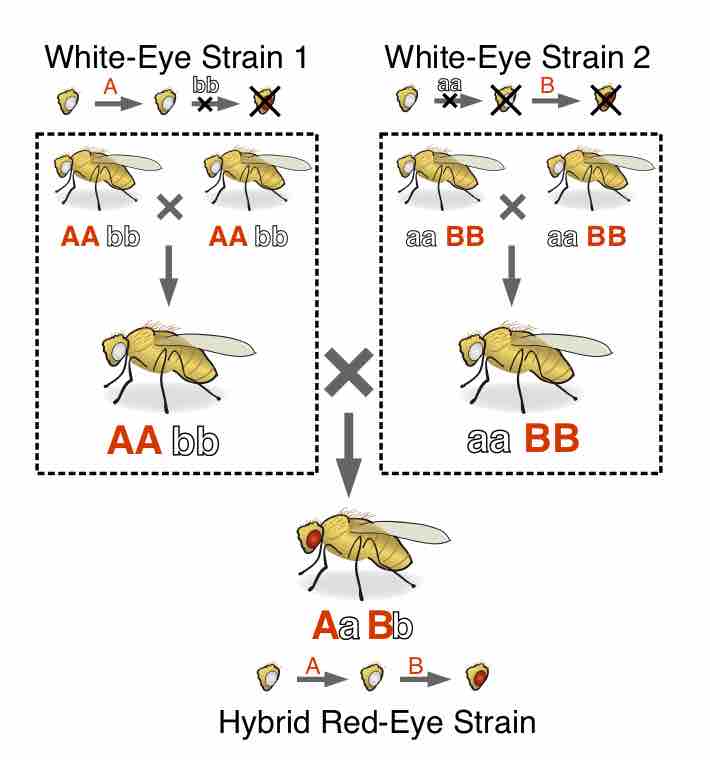
Complementation refers to a relationship between two different strains of an organism which both have homozygous recessive mutations.
Archaea are distinct from bacteria and eukaryotes, but genetic material can be transferred between them and between Archaea themselves.
Molecular cloning permits the replication of a specific DNA sequence in a living microorganism.
A selectable marker is usually a gene that confers resistance to an antibiotic that would otherwise kill the cells.
Mutations are accidental changes in a genomic sequence of DNA; this includes the DNA sequence of a cell's genome or the DNA or RNA sequence.
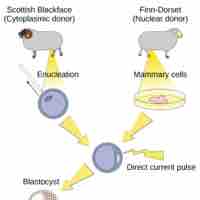
Reproductive cloning, possible through artificially-induced asexual reproduction, is a method used to make a clone of an entire organism.
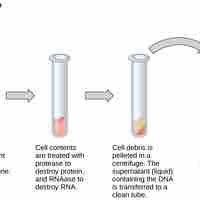
Basic techniques used in genetic material manipulation include extraction, gel electrophoresis, PCR, and blotting methods.
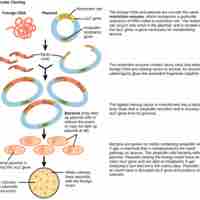
Molecular cloning reproduces the desired regions or fragments of a genome, enabling the manipulation and study of genes.

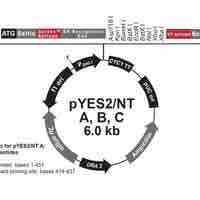
Plasmids can be used as cloning vectors, allowing the insertion of exogenous DNA into a bacterial target.
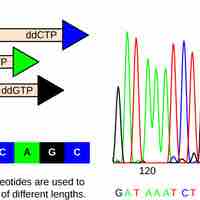
The strategies used for sequencing genomes include the Sanger method, shotgun sequencing, pairwise end, and next-generation sequencing.
Genome annotation is the identification and understanding of the genetic elements of a sequenced genome.

Homology describes the relationship between genes and how they are inherited from ancestors.
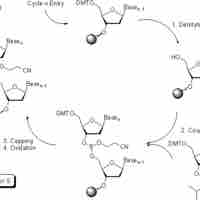
DNA can be synthesized chemically for a number of purposes.
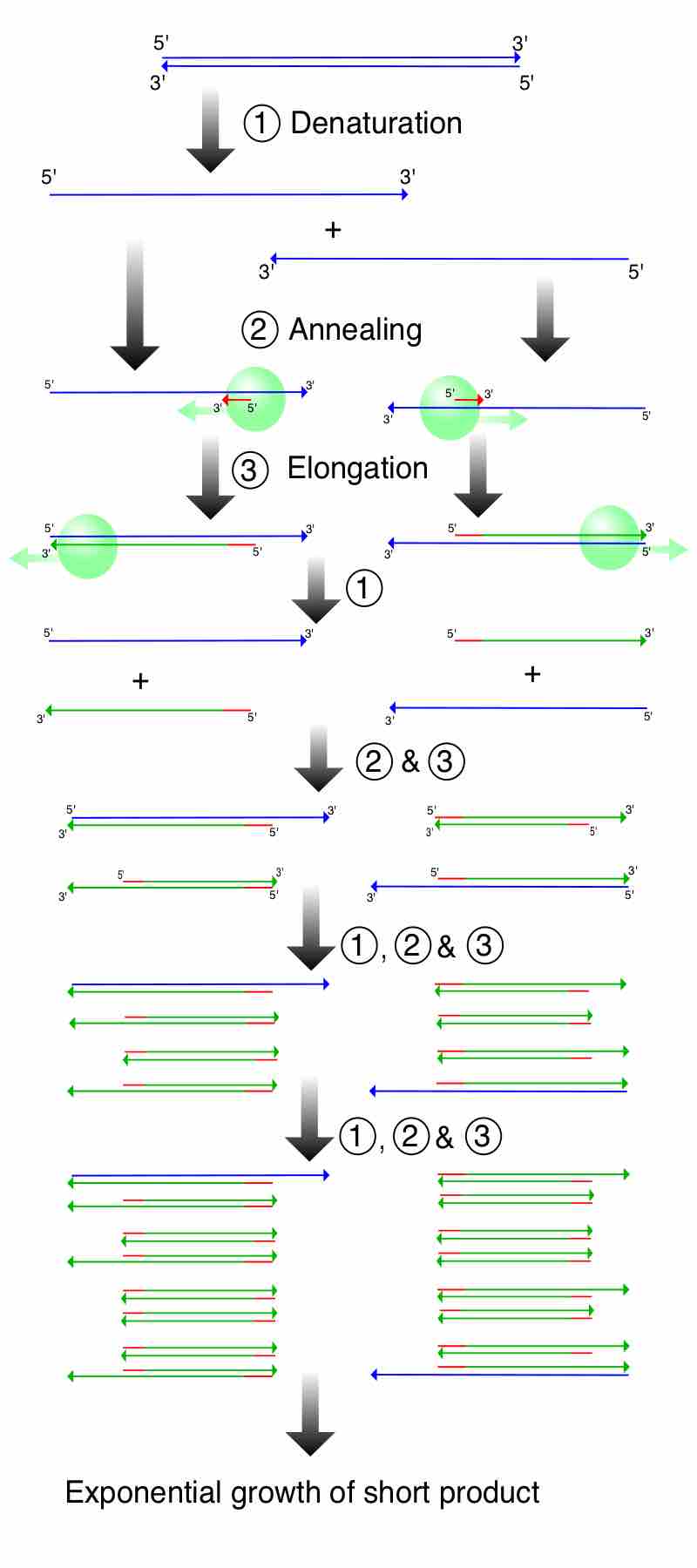
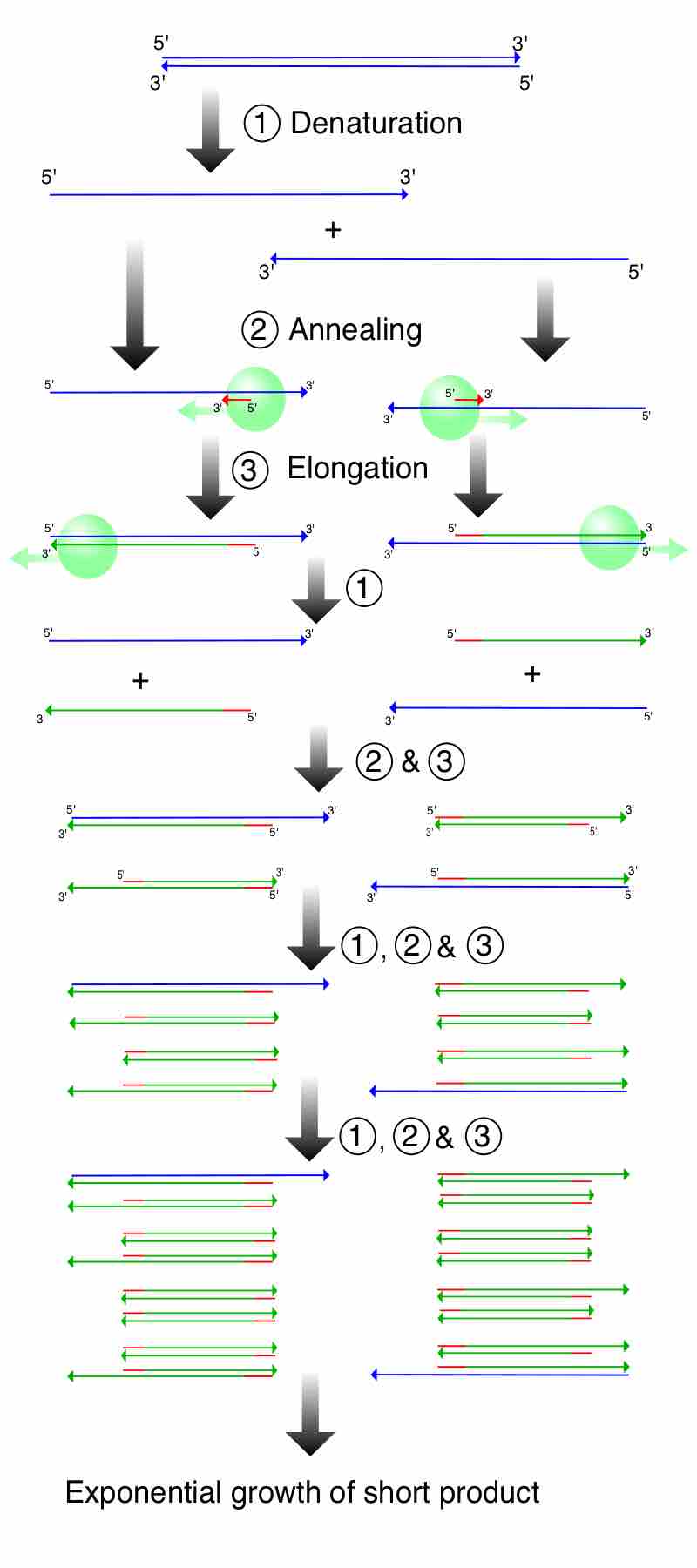
The polymerase chain reaction (PCR) is a method by which DNA is amplified.
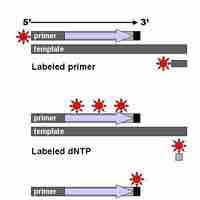
Sanger sequencing is based on the incorporation and detection of labeled ddNTPs as terminal nucleotides in DNA amplification.
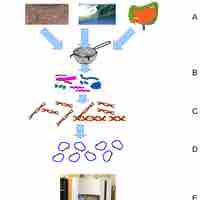
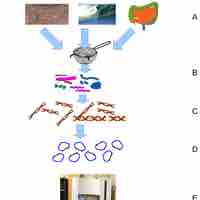
Metagenomics is the study of genetic material derived from environmental samples.
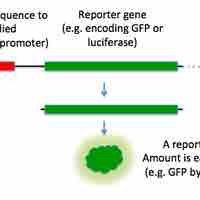
A reporter fusion is the hybrid of a gene or portion of a gene with a tractable marker.
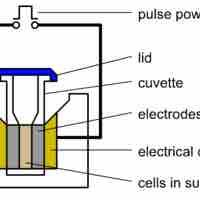
The methods used to get DNA into cells are varied (e.g., transformation, transduction, transfection, and electroporation).
When cloning genomic DNA, the DNA to be cloned is extracted from the organism of interest.
The majority of molecular cloning experiments begin with a laboratory strain of the bacterium E. coli (Escherichia coli) as the host.

An expression vector is generally a plasmid that is used to introduce a specific gene into a target cell.
Enterobacteria phage λ (lambda phage, coliphage λ) is a bacterial virus that infects the bacterial species Escherichia coli.
In molecular biology, a vector is a DNA molecule used as a vehicle to transfer foreign genetic material into another cell.
Gene families are groups of functionally related genes arising from a duplicated gene.

Microbial genomics can be used to create new biofuels.

Genome reduction is the loss of genome size of a species in comparison to its ancestors.
Pathogenenicity islands are discrete genetic loci that encode factors which make a microbe more virulent.
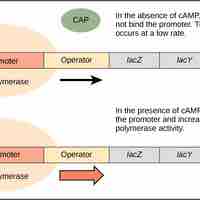
When glucose levels decline in E. coli, catabolite activator protein (CAP) is bound by cAMP to promote transcription of the lac operon.
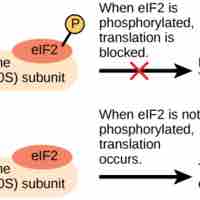
The first step of translation is ribosome assembly, which requires initiation factors.
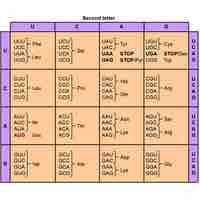
The genetic code is a degenerate, non-overlapping set of 64 codons that encodes for 21 amino acids and 3 stop codons.
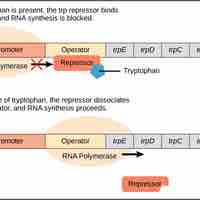
The trp operon is a repressor operon that is either activated or repressed based on the levels of tryptophan in the environment.
The stringent response is a stress response that occurs in bacteria in reaction to amino-acid starvation or other stress conditions.
Repression of anabolic pathways is regulated by altering transcription rates.
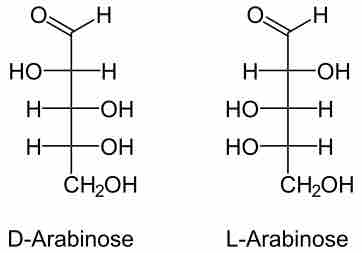
The L-arabinose operon, also called ara operon, encodes enzymes needed for the catabolism of arabinose to xylulose 5-phosphate.
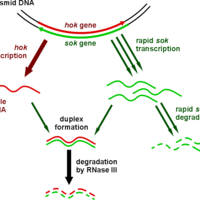
Antisense RNAs are single-stranded RNA molecules that can bind and inhibit specific mRNA translation to protein.

Attenuation is a mechanism utilized by bacteria to regulate unnecessary gene expression.
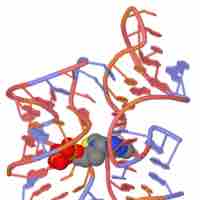
Riboswitches are naturally occurring RNA molecules that can regulate gene expression.
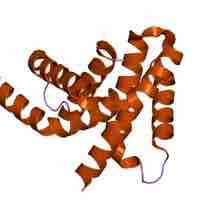
The sigma factor is responsible for proper transcriptional initiation.
Sigma factors are proteins that regulate gene expression that are controlled at various levels, including at the translational level.
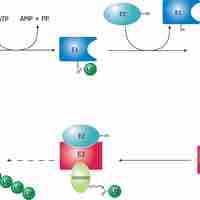
Proteolytic degradation, or proteolysis, is a key factor that controls protein concentration and function.
Small regulatory RNAs are non-coding RNA molecules that play a role in cellular processes such as activation or inhibition processes.
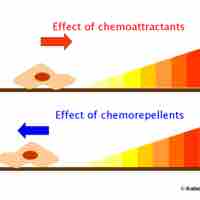
Chemotaxis is the phenomenon whereby bacterial cells direct their movements according to certain chemicals in their environment.
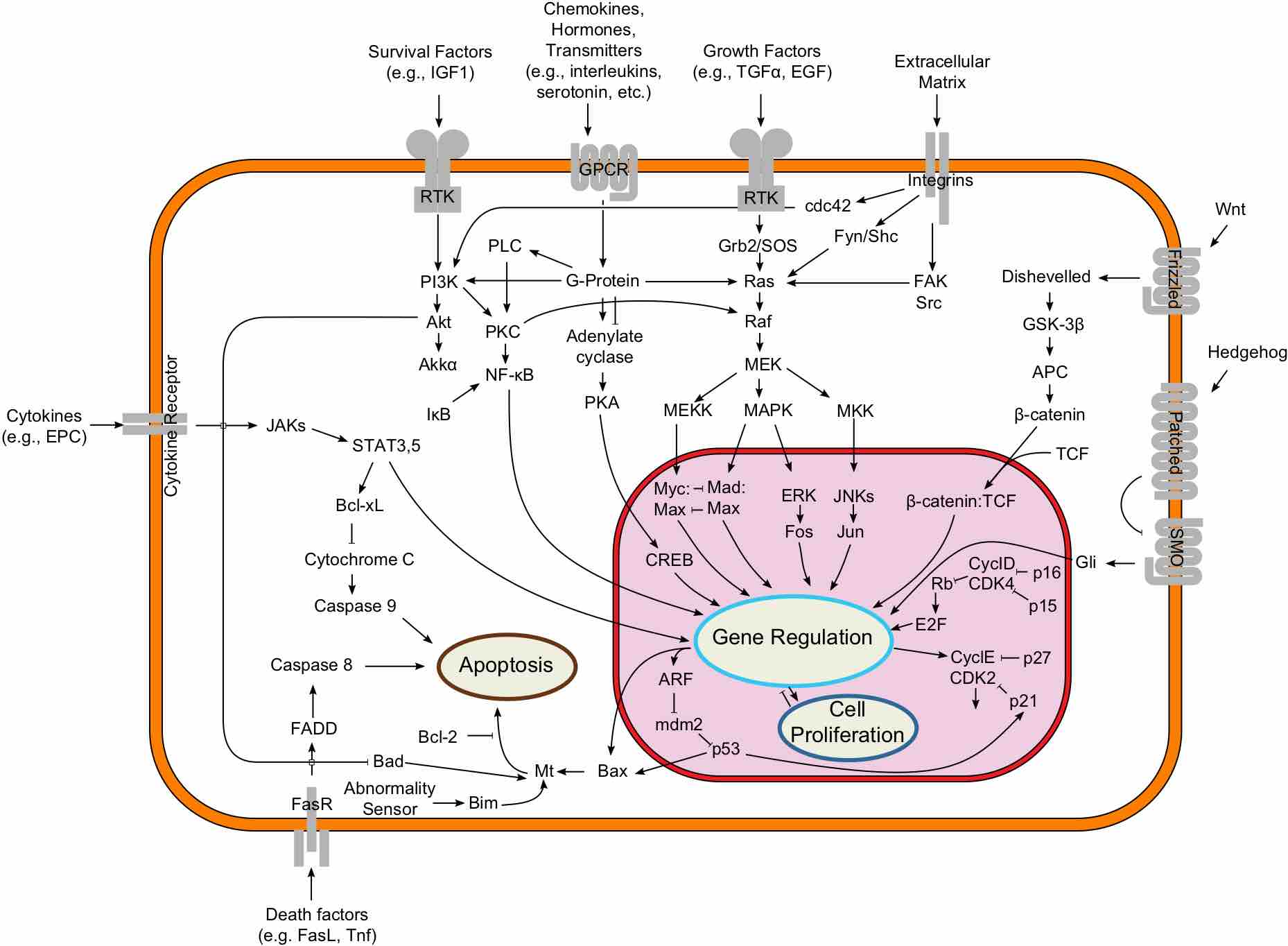
Two-component systems couple mechanism to allow organisms to sense and respond to changes in many different environmental conditions.
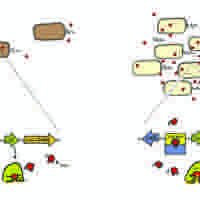
Quorum sensing is a system of stimulus and response correlated to population density.
Transcription and translation in archaea resemble these processes in eukaryotes more than in bacteria.
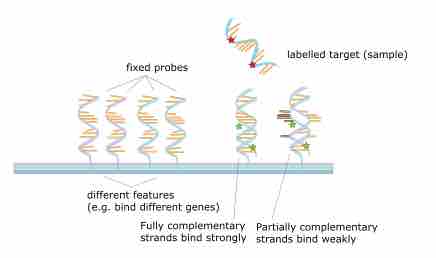
The transcriptome is the set of all RNA molecules, including mRNA, rRNA, tRNA, and other non-coding RNA produced in cells.

Proteomics is the large-scale study of proteins, particularly their structures and functions.

Metabolomics is the scientific study of chemical processes involving metabolites.

Biotechnology is the use of biological techniques and engineered organisms to make products or plants and animals that have desired traits.

Genetic engineering means the manipulation of organisms to make useful products and it has broad applications.
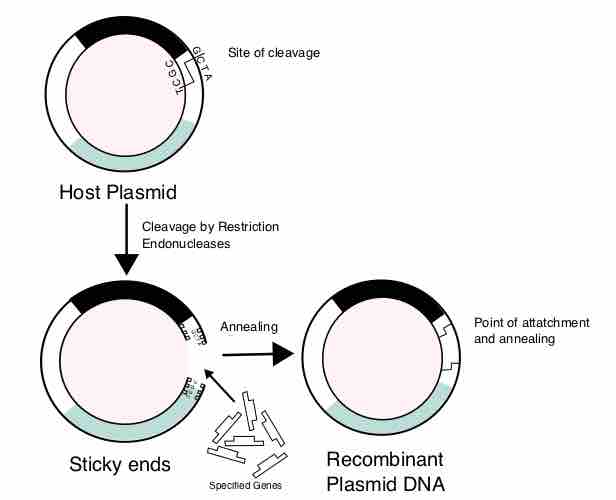
Many practical applications of recombinant DNA are found in human and veterinary medicine, in agriculture, and in bioengineering.
Bacterial genetics can be manipulated to allow for mammalian gene expression systems established in bacteria.
Genetic engineering enables scientists to create plants, animals, and microorganisms by manipulating genes.

Genetic engineering can be used to manufacture new vaccines.

The purpose of genetic engineering in animals is to create animals with special characteristics.
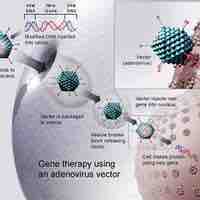
From manipulation of mutant genes to enhanced resistance to disease, biotechnology has allowed advances in medicine.
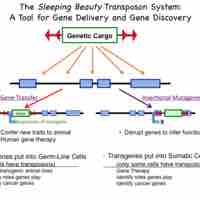
Transposons allow genes to be transferred to a host organism's chromosome, interrupting or modifying the function of a gene.
An insertion site is the position at which a transposable genetic element is integrated.
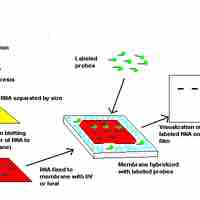
Northern blots allow investigators to determine messenger RNA molecular weight and sample content.
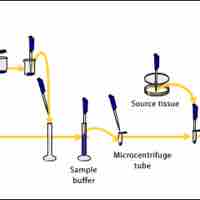
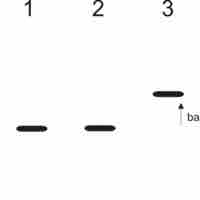
The Western blot technique determines protein molecular weight and measures protein abundance in different samples.
DNA mobility shift assay is a technique for studying gene regulation and determining protein-DNA interactions.
Protein tags are peptide sequences genetically grafted onto a recombinant protein.
Primer extension is used to map the 5' ends of DNA or RNA fragments.
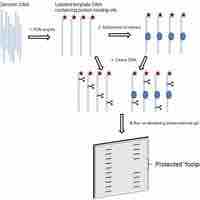
DNA protection or "footprinting" analysis is a powerful technique for identifying the nucleotides involved in a protein-DNA interaction.
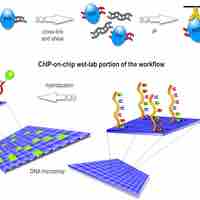
Whole-genome DNA-binding analysis is a powerful tool for analyzing epigenetic modifications and DNA sequences bound to regulatory proteins.

The two-hybrid method detects the interaction of two proteins by their ability to reconstitute the activity of a split transcription factor.
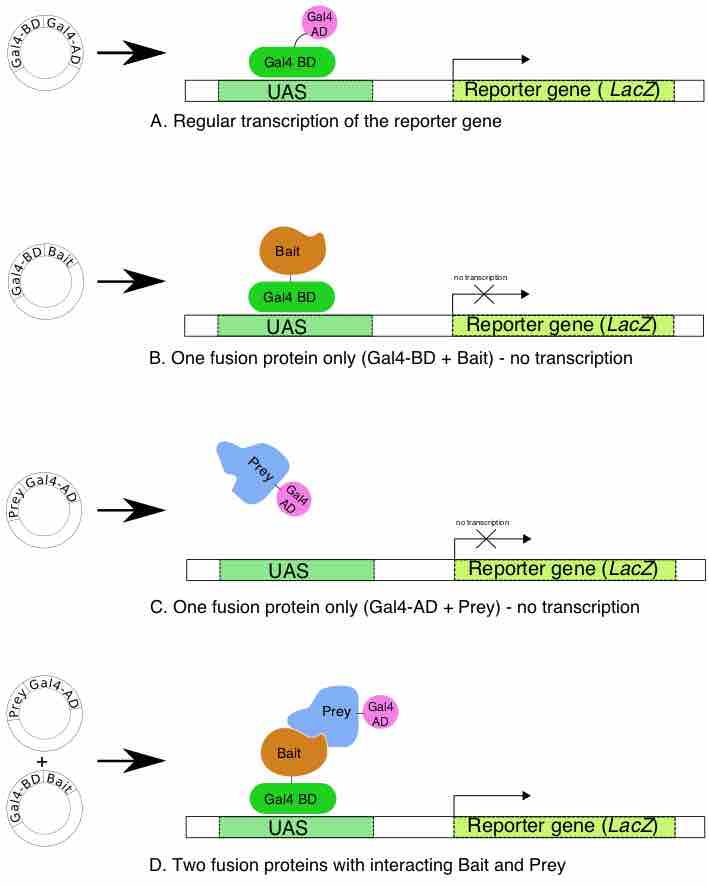
Mapping protein-protein interactions gives us a better understanding of molecular mechanisms inside the cell.
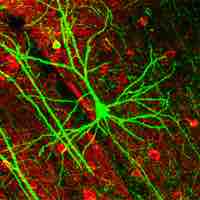
Advanced technology enables tracking cells with light by introducing fluorescent or luminescent reporter genes into the cells' genome.
Multiplex and real-time PCR are molecular techniques designed to amplify nucleic acid sequences in a quantitative manner.
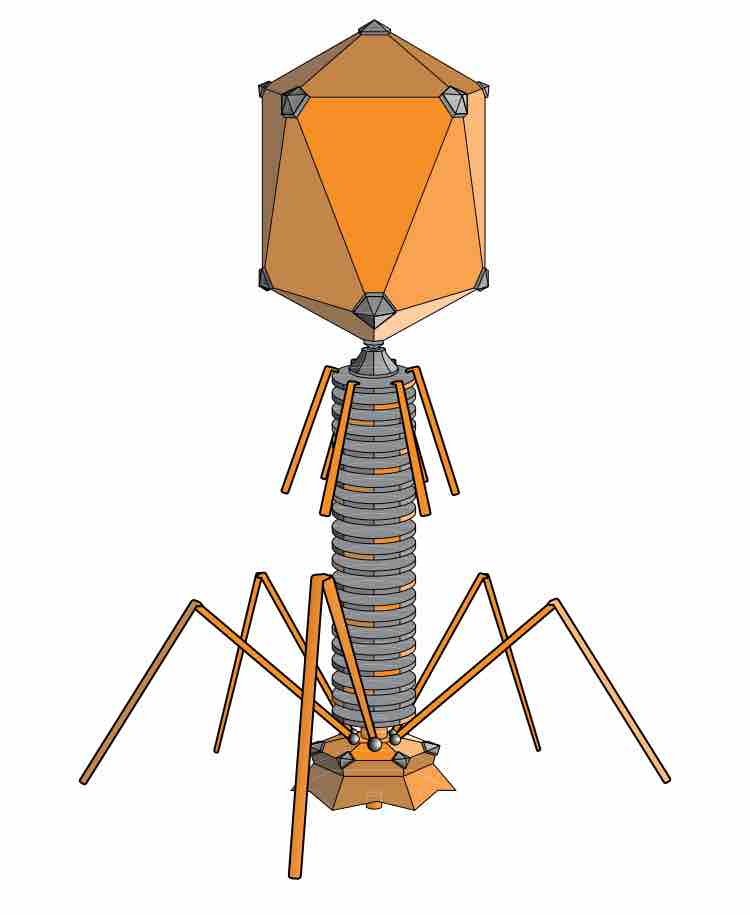
Phage display is a laboratory tool based on cloning DNA sequences into a phage which presents proteins encoded by that DNA on its surface.