Molecular cloning is a set of experimental methods in molecular biology that are used to assemble recombinant DNA molecules and to direct their replication within host organisms. The word cloning in this context refers to the fact that the method involves the replication of a single DNA molecule starting from a single living cell to generate a large population of cells containing identical DNA molecules. Molecular cloning generally uses DNA sequences from two different organisms: the species that is the source of the DNA to be cloned, and the species that will serve as the living host for replication of the recombinant DNA.
For cloning of genomic DNA, the DNA to be cloned is extracted from the organism of interest. Virtually any tissue source can be used (even tissues from extinct animals) as long as the DNA is not extensively degraded. The DNA is then purified using simple methods to remove contaminating proteins (extraction with phenol), RNA (ribonuclease) and smaller molecules (precipitation and/or chromatography). Polymerase chain reaction (PCR) methods are often used for amplification of specific DNA or RNA (by a process known as Reverse-Transcription or RT-PCR) sequences prior to molecular cloning using primers or short DNA sequences specific for the region of interest. DNA for cloning experiments may also be obtained from RNA using reverse transcriptase (complementary DNA or cDNA cloning), or in the form of synthetic DNA (artificial gene synthesis). cDNA cloning is usually used to obtain clones representative of the mRNA population of the cells of interest, while synthetic DNA is used to obtain any precise sequence defined by the designer.
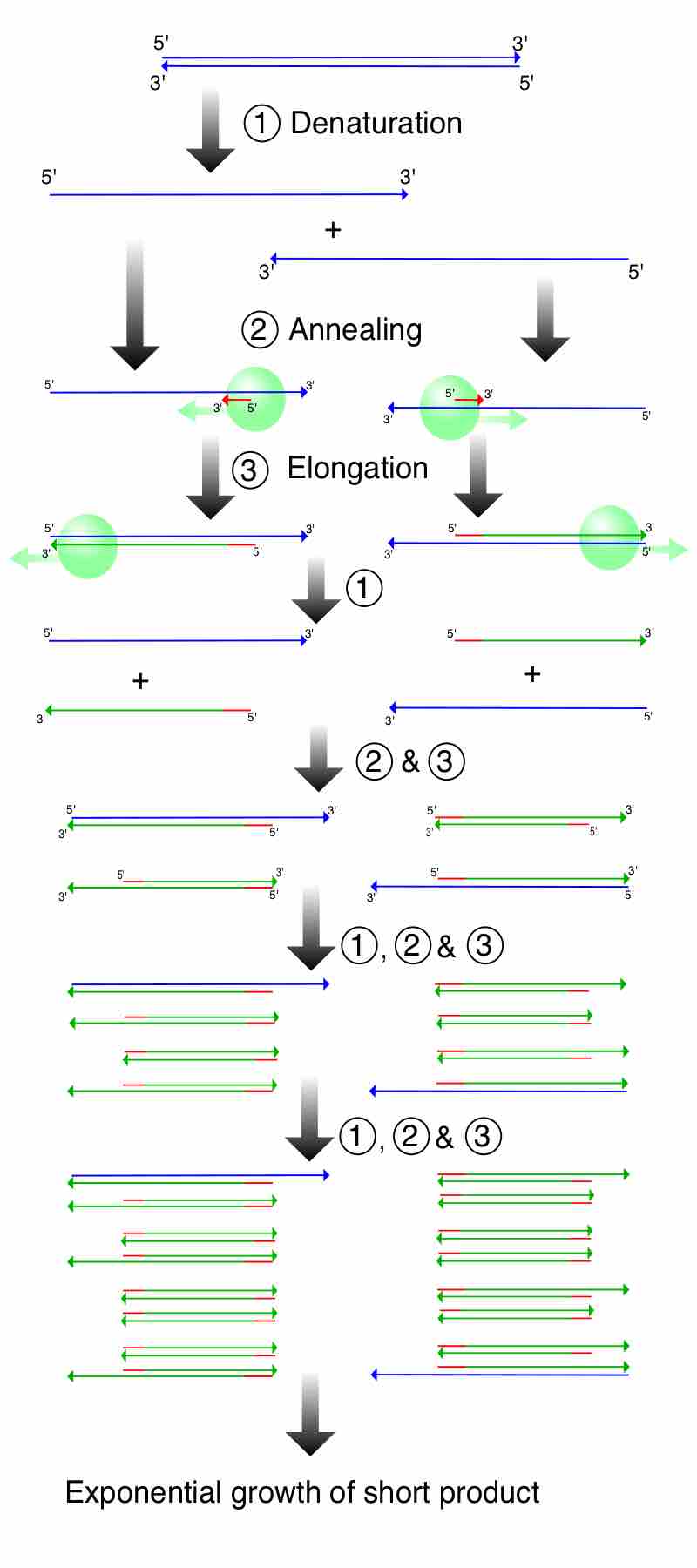
The Steps of PCR
This illustrates a PCR reaction to demonstrate how amplification leads to the exponential growth of a short product flanked by the primers. 1. Denaturing at 96°C. 2. Annealing at 68°C. 3. Elongation at 72°C. The first cycle is complete. The two resulting DNA strands make up the template DNA for the next cycle, thus doubling the amount of DNA duplicated for each new cycle.
Although a very large number of host organisms and molecular cloning vectors are used, the great majority of molecular cloning experiments begin with a laboratory strain of the bacterium E. coli (Escherichia coli) and a plasmid cloning vector. E. coli and plasmid vectors are in common use because they are technically sophisticated, versatile, widely available and offer rapid growth of recombinant organisms with minimal equipment. If the DNA to be cloned is exceptionally large (hundreds of thousands to millions of base pairs), then a bacterial artificial chromosome or yeast artificial chromosome vector is often chosen.
The cloning vector is treated with a restriction endonuclease to cleave the DNA at the site where foreign DNA will be inserted. The restriction enzyme is chosen to generate a configuration at the cleavage site that is compatible with that at the ends of the foreign DNA. Typically, this is done by cleaving the vector DNA and foreign DNA with the same restriction enzyme. Most modern vectors contain a variety of convenient cleavage sites that are unique within the vector molecule (so that the vector can only be cleaved at a single site) and are not located within the gene of interest to be cloned.
The creation of recombinant DNA is in many ways the simplest step of the molecular cloning process. DNA prepared from the vector and foreign source are treated with restriction enzymes to generate fragments with ends capable of being linked to those of the vector and they are simply mixed together at appropriate concentrations and exposed to an enzyme (DNA ligase) that covalently links the ends together. This joining reaction is often termed ligation. The resulting DNA mixture containing randomly joined ends is then ready for introduction into the host organism for amplification (a process known as transformation). In mammalian cell culture, the analogous process of introducing DNA into cells is commonly known as transfection. Both transformation and transfection usually require preparation of the cells through a special growth regimen and chemical treatment process that will vary with the specific species and cell types that are used. Whichever method is used, the introduction of recombinant DNA into the chosen host organism is usually a low efficiency process; that is, only a small fraction of the cells will actually take up DNA. When bacterial cells are used as host organisms, the selectable marker is usually a gene that confers resistance to an antibiotic, typically ampicillin, that would otherwise kill the cells. Cells harboring the cloning vector will survive when exposed to the antibiotic, while those that have failed to take up cloning vector will die. The former can therefore be amplified and screened for the presence of the gene of interest in the cloning vector by restriction digest analysis.