In molecular genetics, an open reading frame (ORF) is the part of a reading frame that contains no stop codons. The transcription termination pause site is located after the ORF, beyond the translation stop codon, because if transcription were to cease before the stop codon, an incomplete protein would be made during translation.
Normally, inserts which interrupt the reading frame of a subsequent region after the start codon cause frameshift mutation of the sequence and dislocate the sequences for stop codons.
Open reading frames are used as one piece of evidence to assist in gene prediction. Long ORFs are often used, along with other evidence, to initially identify candidate protein coding regions in a DNA sequence . The presence of an ORF does not necessarily mean that the region is ever translated. For example, in a randomly generated DNA sequence with an equal percentage of each nucleotide, a stop-codon would be expected once every 21 codons. A simple gene prediction algorithm for prokaryotes might look for a start codon followed by an open reading frame that is long enough to encode a typical protein, where the codon usage of that region matches the frequency characteristic for the given organism's coding regions. Even a long open reading frame by itself is not conclusive evidence for the presence of a gene.
Open Reading Frames
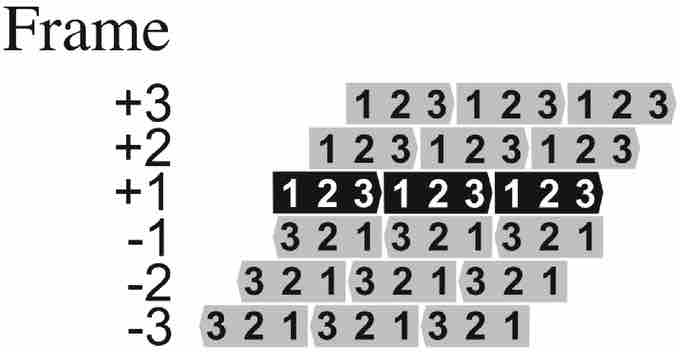
Frame +1 is the ORF predicted in the database to encode a protein. +2 and +3 are the other two potential ORFs in the same strand and -1, -2, and -3 are the three potential ORFs in the antisense strand.
If a portion of a genome has been sequenced (e.g. 5'-ATCTAAAATGGGTGCC-3'), ORFs can be located by examining each of the three possible reading frames on each strand. In this sequence two out of three possible reading frames are entirely open, meaning that they do not contain a stop codon:
...A TCT AAA ATG GGT GCC...
...AT CTA AAA TGG GTG CC...
...ATC TAA AAT GGG TGC C...
Possible stop codons in DNA are "TGA", "TAA", and "TAG". Thus, the last reading frame in this example contains a stop codon (TAA), unlike the first two.
Bacterial genomes display variation in size, even among strains of the same species. These microorganisms have very little noncoding or repetitive DNA, as the variation in their genome size usually reflects differences in gene repertoire. Some species, particularly bacterial parasites and symbionts, have undergone massive genome reduction and simply contain a subset of the genes present in their ancestors.
However, in free-living bacteria, such gene loss cannot explain the observed disparities in genome size because ancestral genomes would have had to contain improbably large numbers of genes. Surprisingly, a substantial fraction of the difference in gene contents in free-living bacteria is due to the presence of ORFans, that is, open reading frames (ORFs) that have no known homologs and are consequently of no known function.
The high numbers of ORFans in bacterial genomes indicate that, with the exception of those species with highly reduced genomes, much of the observed diversity in gene inventories does not result from either the loss of ancestral genes or the transfer from well-characterized organisms (processes that result in a patchy distribution of orthologs but not in unique genes) or from recent duplications (which would likely yield homologs within the same or closely related genome).