Repression of anabolic pathways is regulated by altering transcription rates. Transcriptional regulation is the change in gene expression levels by altering transcription rates.
Regulation of transcription controls when transcription occurs and how much RNA is created. Transcription of a gene by RNA polymerase can be regulated by at least five mechanisms:
- Specificity factors alter the specificity of RNA polymerase for a given promoter or set of promoters, making it more or less likely to bind to them (i.e. sigma factors used in prokaryotic transcription).
- Repressors bind to non-coding sequences on the DNA strand that are close to or overlapping the promoter region, impeding RNA polymerase's progress along the strand, thus impeding the expression of the gene.
- General transcription factors position RNA polymerase at the start of a protein-coding sequence and then release the polymerase to transcribe the mRNA.
- Activators enhance the interaction between RNA polymerase and a particular promoter, encouraging the expression of the gene. Activators do this by increasing the attraction of RNA polymerase for the promoter, through interactions with subunits of the RNA polymerase or indirectly by changing the structure of the DNA.
- Enhancers are sites on the DNA helix that are bound to by activators in order to loop the DNA bringing a specific promoter to the initiation complex.
Regulatory protein is a term used in genetics to describe a protein involved in regulating gene expression. Such proteins are usually bound to a DNA binding site which is sometimes located near the promoter although this is not always the case. Sites of DNA sequences where regulatory proteins bind are called enhancer sequences. Regulatory proteins are often needed to be bound to a regulatory binding site to switch a gene on (activator) or to shut off a gene (repressor). Generally, as the organism grows more sophisticated, its cellular protein regulation becomes more complicated and, indeed, some human genes can be controlled by many activators and repressors working together.
In prokaryotes, regulation of transcription is needed for the cell to quickly adapt to the ever-changing outer environment. The presence or the quantity and type of nutrients determines which genes are expressed; in order to do that, genes must be regulated in some fashion. In prokaryotes, repressors bind to regions called operators that are generally located downstream from and near the promoter (normally part of the transcript). Activators bind to the upstream portion of the promoter, such as the CAP region (completely upstream from the transcript). A combination of activators, repressors and rarely enhancers (in prokaryotes) determines whether a gene is transcribed.
Gene regulation can be summarized as how genes respond: inducible systems or repressible systems. An inducible system is off unless there is the presence of some molecule (called an inducer) that allows for gene expression. The molecule is said to "induce expression. " The manner in which this happens is dependent on the control mechanisms as well as differences between prokaryotic and eukaryotic cells. A repressible system is on except in the presence of some molecule (called a corepressor) that suppresses gene expression. The molecule is said to "repress expression. " The manner in which this happens is dependent on the control mechanisms as well as differences between prokaryotic and eukaryotic cells.
For example, when E. coli bacteria are subjected to heat stress, the σ32 subunit of its RNA polymerase changes in such a way that the enzyme binds to a specialized set of promoters that precede genes for heat-shock response proteins.
Another example is when a cell contains a surplus amount of the amino acid tryptophan, the acid binds to a specialized repressor protein (tryptophan repressor) . The binding changes the structural conformity of the repressor such that it binds to the operator region for the operon that synthesizes tryptophan, preventing their expression and thus suspending production. This is a form of negative feedback.
Trp Repressor Protein
Here is a diagram of the Trp repressor protein.
In bacteria, the lac repressor protein blocks the synthesis of enzymes that digest lactose when there is no lactose to feed on. When lactose is present, it binds to the repressor, causing it to detach from the DNA strand.
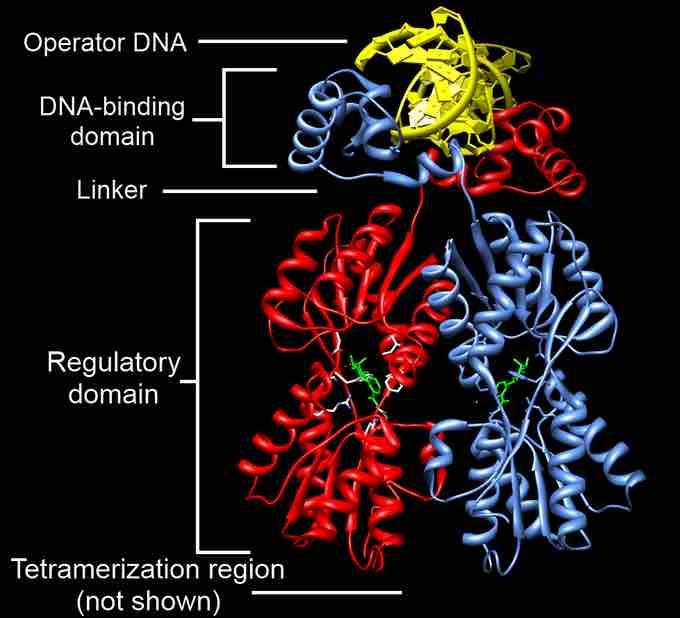
Annotated Crystal Structure of Dimeric LacI
Annotated crystal structure of dimeric LacI. Two monomers (of four total) cooperate to bind each DNA operator sequence. Monomers (red and blue) contain DNA binding and core domains (labeled) which are connected by a linker (labeled). The C-terminal tetramerization helix is not shown. The repressor is shown in complex with operator DNA (gold) and ONPF (green), an anti-inducer ligand (i.e. a stabilizer of DNA binding).