The polymerase chain reaction (PCR) is a biochemical technology in molecular biology used to amplify a single, or a few copies, of a piece of DNA across several orders of magnitude, generating thousands to millions of copies of a particular DNA sequence.
Applications
Developed in 1983 by Kary Mullis, PCR is now a common and often indispensable technique used in medical and biological research labs for a variety of applications including the following:
- DNA cloning for sequencing; DNA-based phylogeny, or functional analysis of genes
- The diagnosis of hereditary diseases
- The identification of genetic fingerprints (used in forensic sciences and paternity testing)
- The detection and diagnosis of infectious diseases
The method relies on thermal cycling, consisting of cycles of repeated heating and cooling of the reaction for DNA melting and enzymatic replication of the DNA. Primers (short DNA fragments) containing sequences complementary to the target region, along with a DNA polymerase (after which the method is named) are key components to enable selective and repeated amplification. As PCR progresses, the DNA generated is itself used as a template for replication, setting in motion a chain reaction in which the DNA template is exponentially amplified. PCR can be extensively modified to perform a wide array of genetic manipulations.
Components
PCR is used to amplify a specific region of a DNA strand (the DNA target). Most PCR methods typically amplify DNA fragments of up to ~10 kilo base pairs (kb), although some techniques allow for amplification of fragments up to 40 kb in size. The reaction produces a limited amount of final amplified product that is governed by the available reagents in the reaction, and the feedback-inhibition of the reaction products. A basic PCR set up requires the following components and reagents:
- DNA template that contains the DNA region (target) to be amplified
- Two primers that are complementary to the 3' (three prime) ends of each of the sense and anti-sense strand of the DNA target
- Taq polymerase or another DNA polymerase with a temperature optimum at around 70 °C
- Deoxynucleoside triphosphates (dNTPs; nucleotides containing triphosphate groups), the building-blocks from which the DNA polymerase synthesizes a new DNA strand
- Buffer solution, providing a suitable chemical environment for optimum activity and stability of the DNA polymerase.Divalent cations, magnesium or manganese ions; generally Mg2+
- Monovalent cation potassium ions
Typically, PCR consists of a series of 20-40 repeated temperature changes, called cycles, with each cycle commonly consisting of two to three discrete temperature steps, usually three. The temperatures used, and the length of time they are applied in each cycle, depend on a variety of parameters. These include the enzyme used for DNA synthesis, the concentration of divalent ions and dNTPs in the reaction, and the melting temperature (Tm) of the primers.
Steps
The following are the steps of PCR :
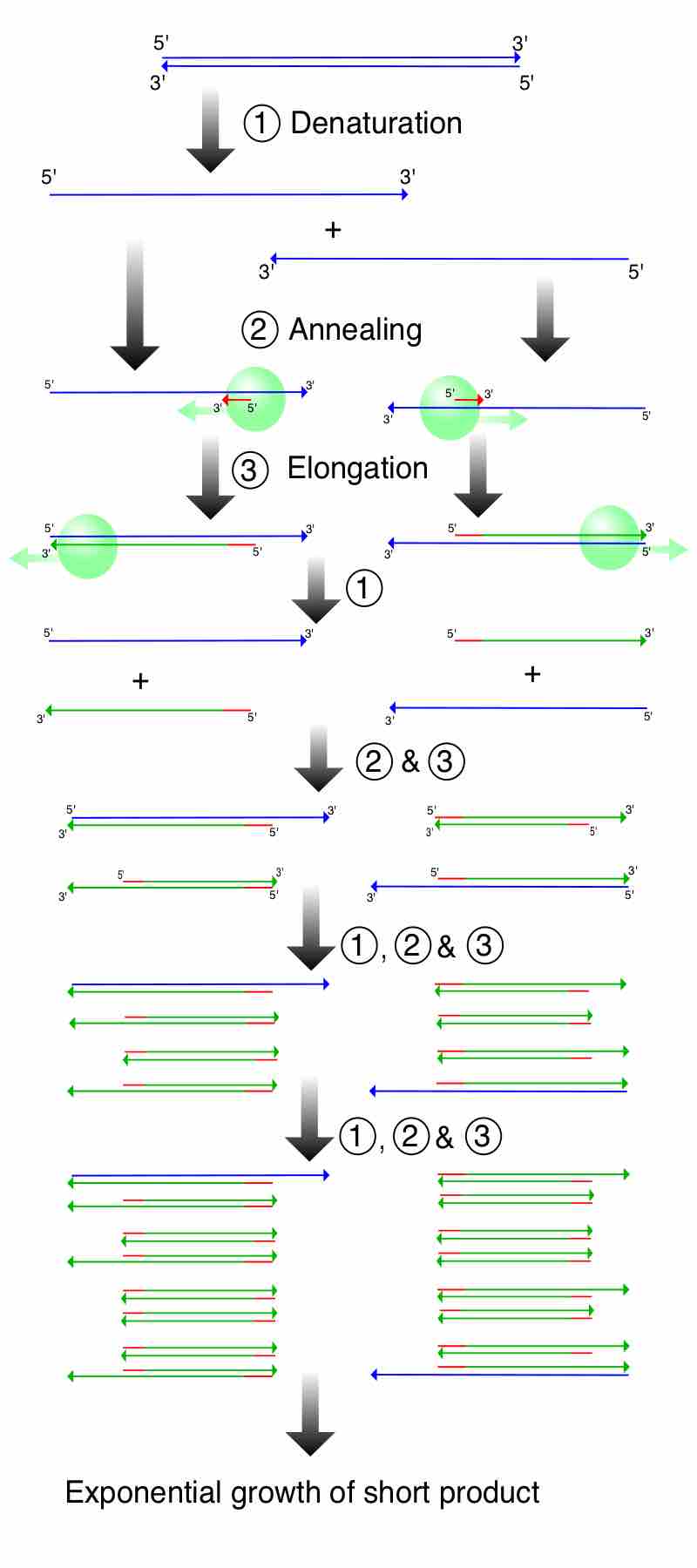
The Steps of PCR
This illustrates a PCR reaction to demonstrate how amplification leads to the exponential growth of a short product flanked by the primers. 1. Denaturing at 96°C. 2. Annealing at 68°C. 3. Elongation at 72°C. The first cycle is complete. The two resulting DNA strands make up the template DNA for the next cycle, thus doubling the amount of DNA duplicated for each new cycle.
- Denaturation step: This step is the first regular cycling event and consists of heating the reaction to 94-98 °C. It causes DNA melting of the DNA template by disrupting the hydrogen bonds between complementary bases, yielding single-stranded DNA molecules.
- Annealing step: The reaction temperature is lowered to 50-65 °C for 20-40 seconds allowing annealing of the primers to the single-stranded DNA template.
- Extension/elongation step: The temperature at this step depends on the DNA polymerase used; Taq polymerase has its optimum activity temperature at 75-80 °C, and commonly a temperature of 72 °C is used with this enzyme. At this step the DNA polymerase synthesizes a new DNA strand complementary to the DNA template strand by adding dNTPs that are complementary to the template in 5' to 3' direction, condensing the 5'-phosphate group of the dNTPs with the 3'-hydroxyl group at the end of the nascent (extending) DNA strand.
After elongation, the cycle goes back to step one, usually for 20-40 cycles. Under optimum conditions (i.e., if there are no limitations due to limiting substrates or reagents) at each extension step, the amount of DNA target is doubled, leading to exponential (geometric) amplification of the specific DNA fragment.