A transposable element (TE) is a DNA sequence that can change its relative position (self-transpose) within the genome of a single cell. The mechanism of transposition can be either "copy and paste" or "cut and paste. " Transposition can create phenotypically significant mutations and alter the cell's genome size. Barbara McClintock's discovery of these jumping genes early in her career earned her a Nobel prize in 1983.
Transposons in bacteria usually carry an additional gene for function other than transposition---often for antibiotic resistance. In bacteria, transposons can jump from chromosomal DNA to plasmid DNA and back, allowing for the transfer and permanent addition of genes such as those encoding antibiotic resistance (multi-antibiotic resistant bacterial strains can be generated in this way). When the transposable elements lack additional genes, they are known as insertion sequences. Transposons are semi-parasitic DNA sequences that can replicate and spread through the host's genome. They can be harnessed as a genetic tool for analysis of gene and protein function. The use of transposons is well-developed in Drosophila (in which P elements are most commonly used) and in Thale cress (Arabidopsis thaliana) and bacteria such as Escherichia coli (E. coli ).
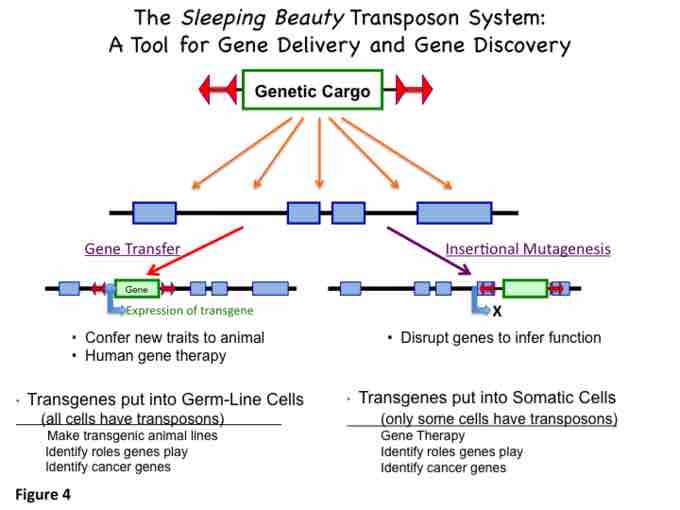
Synthetic DNA transposon system are constructed to introduce precisely defined DNA sequences into the chromosomes of vertebrate animals for the purposes of introducing new traits and to discover new genes and their functions (e.g. by establishing a loss-of-function phenotype or gene inactivation). Transposition is a precise process in which a defined DNA segment is excised from one DNA molecule and moved to another site in the same or different DNA molecule or genome.
Transposon System
The sleeping beauty transposon system applications.
Insertional inactivation is a technique used in recombinant DNA engineering where a plasmid (such as pBR322) is used to disable the expression of a gene. A gene is inactivated by inserting a fragment of DNA into the middle of its coding sequence. Any future products from the inactivated gene will not work because of the extra codes added to it. An example is the use of pBR322, which has genes that respectively encode polypeptides that confer resistance to ampicillin and tetracyclin antibiotics. As a result, when a genetic region is interrupted by integration of pBR322, the gene function is lost but new gene function (resistance to specific antibiotics) is gained. An alternative strategy for insertional mutagenesis has been used in vertebrate animals to find genes that cause cancer. In this case a transposon, e.g. Sleeping Beauty, is designed to interrupt a gene in such a way that it causes maximal genetic havoc. Specifically, the transposon contains signals to truncate expression of an interrupted gene at the site of the insertion and then restart expression of a second truncated gene. This method has been used to identify oncogenes.