An insertion sequence (also known as an IS, an insertion sequence element, or an IS element) is a short DNA sequence that acts as a simple transposable element.
Insertion sequences have two major characteristics: they are small relative to other transposable elements (generally around 700 to 2500 bp in length) and only code for proteins implicated in the transposition activity (they are thus different from other transposons, which also carry accessory genes such as antibiotic-resistance genes).
These proteins are usually the transposase which catalyse the enzymatic reaction allowing the IS to move, and also one regulatory protein which either stimulates or inhibits the transposition activity. The coding region in an insertion sequence is usually flanked by inverted repeats. For example, the well-known IS911 (1250 bp) is flanked by two 36bp inverted repeat extremities and the coding region has two genes partially overlapping orfA and orfAB, coding the transposase (OrfAB) and a regulatory protein (OrfA).
A particular insertion sequence may be named according to the form ISn, where n is a number (e.g. IS1, IS2, IS3, IS10, IS50, IS911, IS26, etc.); this is not the only naming scheme used, however. Although insertion sequences are usually discussed in the context of prokaryotic genomes, certain eukaryotic DNA sequences belonging to the family of Tc1/mariner transposable elements may be considered to be insertion sequences.
In addition to occurring autonomously, insertion sequences may also occur as parts of composite transposons; in a composite transposon, two insertion sequences flank one or more accessory genes, such as an antibiotic-resistance gene (e.g. Tn10, Tn5). Nevertheless, there exist another sort of transposons, called unit transposons, that do not carry insertion sequences at their extremities (e.g. Tn7). A complex transposon does not rely on flanking insertion sequences for resolvase. The resolvase is part of the tns genome and cuts at flanking inverted repeats.
Although several methods are available for locating ISs in microbial genomes, they are either labor intensive or inefficient. These include Southern hybridization, inverse Polymerase Chain Reaction (iPCR), and most recently, vectorette PCR to identify and map the genomic positions of the insertion sequences.
Southern hybridization is rather time-consuming and requires additional procedures for localizing ISs. Inverse PCR, a commonly-used PCR method for recovering unknown flanking sequences of a known target sequence, uses a library of circularized chromosomal DNA fragments as a template and two outward primers located in each end of the known fragment for amplification . However, when a target sequence has multiple genomic locations, the variously-sized DNA circles formed are difficult to amplify simultaneously. Also, the length of each restriction DNA fragment containing a target sequence must be determined by Southern hybridization followed by sub-genomic fractioning before intramolecular ligation and PCR amplification. These difficulties render Southern hybridization and iPCR impractical as techniques for quickly surveying repetitive elements in genomes.
Naegleria fowleri
Antibody detection (green) of Naegleria fowleri, the organism responsible for Primary amoebic meningoencephalitis (PAM).
Vectorette PCR (vPCR) is another method used to amplify unknown sequences flanking a characterized DNA fragment. It involves cutting genomic DNAs with a restriction enzyme, ligating vectorettes to the ends, and amplifying the flanking sequences of a known sequence using primers derived from the known sequence along with a vectorette primer.
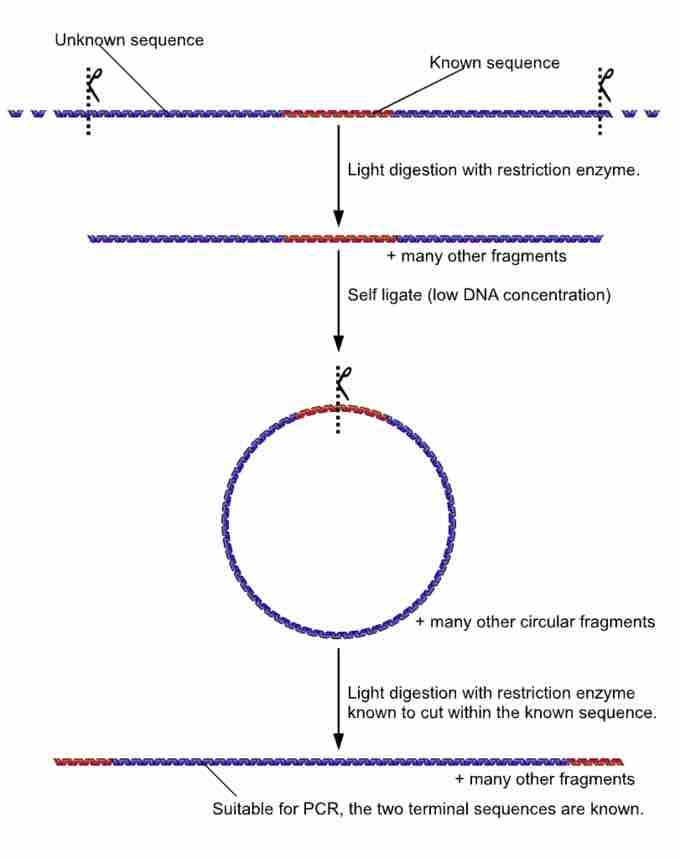
Inverse PCR
Summary of iPCR process