The Genetic Code: Nucleotide sequences prescribe the amino acids
The genetic code is the relationship between DNA base sequences and the amino acid sequence in proteins. Features of the genetic code include:
- Amino acids are encoded by three nucleotides.
- It is non-overlapping.
- It is degenerate.
There are 21 genetically-encoded amino acids universally found in the species from all three domains of life. ( There is a 22nd genetically-encooded amino acid, Pyl, but so far it has only been found in a handful of Archaea and Bacteria species.) Yet there are only four different nucleotides in DNA or RNA, so a minimum of three nucleotides are needed to code each of the 21 (or 22) amino acids . The set of three nucleotides that codes for a single amino acid is known as a codon. There are 64 codons in total, 61 that encode amino acids and 3 that code for chain termination. Two of the codons for chain termination can, under certain circumstances, instead code for amino acids.
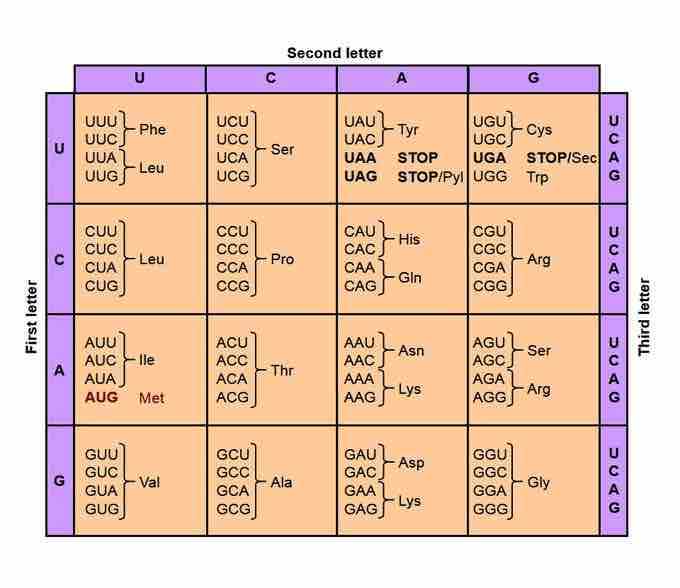
Genetic Code Table.
A codon is made of three nucleotides. Consequently there are 43 (=64) different codons. The 64 codons encode 22 different amino acids and three termination codons, also called stop codons.
Degeneracy is the redundancy of the genetic code. The genetic code has redundancy, but no ambiguity. For example, although codons GAA and GAG both specify glutamic acid (redundancy), neither of them specifies any other amino acid (no ambiguity). The codons encoding one amino acid may differ in any of their three positions. For example, the amino acid glutamic acid is specified by GAA and GAG codons (difference in the third position); the amino acid leucine is specified by UUA, UUG, CUU, CUC, CUA, CUG codons (difference in the first or third position); while the amino acid serine is specified by UCA, UCG, UCC, UCU, AGU, AGC (difference in the first, second or third position). These properties of the genetic code make it more fault-tolerant for point mutations.
Origin of transcription on prokaryotic organisms
Prokaryotes are mostly single-celled organisms that, by definition, lack membrane-bound nuclei and other organelles. The central region of the cell in which prokaryotic DNA resides is called the nucleoid region. Bacterial and Archaeal chromosomes are covalently-closed circles that are not as extensively compacted as eukaryotic chromosomes, but are compacted nonetheless as the diameter of a typical prokaryotic chromosome is larger than the diameter of a typical prokaryotic cell. Additionally, prokaryotes often have abundant plasmids, which are shorter, circular DNA molecules that may only contain one or a few genes and often carry traits such as antibiotic resistance.
Transcription in prokaryotes (as in eukaryotes) requires the DNA double helix to partially unwind in the region of RNA synthesis. The region of unwinding is called a transcription bubble. Transcription always proceeds from the same DNA strand for each gene, which is called the template strand. The RNA product is complementary to the template strand and is almost identical to the other (non-template) DNA strand, called the sense or coding strand. The only difference is that in RNA all of the T nucleotides are replaced with U nucleotides.
The nucleotide on the DNA template strand that corresponds to the site from which the first 5' RNA nucleotide is transcribed is called the +1 nucleotide, or the initiation site. Nucleotides preceding, or 5' to, the template strand initiation site are given negative numbers and are designated upstream. Conversely, nucleotides following, or 3' to, the template strand initiation site are denoted with "+" numbering and are called downstream nucleotides.