Virtually any microbe can trigger an antibody response. Successful recognition and eradication of a variety of microbes requires diversity among antibodies (i.e., a varying amino acid composition allowing them to interact with many different antigens). Humans generate an estimated 10 billion different antibodies, each capable of binding a distinct antigen epitope. Although a single individual contains a wide arsenal of antibodies, the number of genes available to make these proteins is limited by the size of the human genome. Several complex genetic mechanisms have evolved allowing vertebrate B cells to generate a diverse pool of antibodies from a relatively small number of antibody genes.
Domain Variability
The region (locus) of a chromosome that encodes an antibody is large and contains several distinct genes for each antibody domain— in humans the locus containing heavy chain genes (IGH) is found on chromosome 14, and the loci containing lambda and kappa light chain genes (IGL and IGK) are found on chromosomes 22 and 2. One of these domains, the variable domain, is present in the heavy and light chain of every antibody, but is differentiated in antibodies generated from distinct B cells. Differences between the variable domains are located on three loops known as hypervariable regions (HV-1, HV-2 and HV-3) or complementarity determining regions (CDR1, CDR2 and CDR3). CDRs are supported within the variable domains by conserved framework regions. The heavy chain locus contains about 65 different variable domain genes, all with differing CDRs. Combining these genes with an assortment of genes for other antibody domains generates a large cavalry of antibodies (i.e., a high degree of variability). This combination is called V(D)J recombination, discussed below.
Somatic Hypermutation and Affinity Maturation
Following activation with antigen, B cells proliferate rapidly. In these rapidly dividing cells, the genes encoding the variable domains of the heavy and light chains undergo a high rate of point mutation by a process called somatic hypermutation (SHM). SHM results in approximately one nucleotide change per variable gene, per cell division. As a consequence, any daughter B cells acquire slight amino acid differences in the variable domains of their antibody chains, increasing the diversity of the antibody pool and impacting the antibody's antigen-binding affinity. Some point mutations result in the production of antibodies having a weaker interaction (low affinity) with their antigen than the original antibody, and some generate antibodies with a stronger interaction (high affinity). B cells that express high affinity antibodies on their surface receive a strong survival signal during interactions with other cells, whereas those with low affinity antibodies do not, and will die by apoptosis. Thus, B cells expressing antibodies with a higher affinity for the antigen outcompete those with weaker affinities for function and survival. The process of generating antibodies with increased binding affinities is called affinity maturation; this occurs in mature B cells after V(D)J recombination and is dependent on help from helper T cells.
Mechanism of Class Switching
Isotype or class switching is a biological process occurring after activation of the B cell, allowing the cell to produce different classes of antibodies (IgA, IgE, or IgG) . The different classes of antibody (and thus effector functions) are defined by the constant (C) regions of the immunoglobulin heavy chain. Initially, naïve B cells express only cell-surface IgM and IgD with identical antigen binding regions. Each isotype is adapted for a distinct function. After activation, accordingly, an antibody with a IgG, IgA, or IgE effector function might be summoned to effectively eliminate an antigen. Class switching allows different daughter cells from the same activated B cell to produce antibodies of different isotypes. Only the constant region of the antibody heavy chain changes during class switching; the variable regions, and therefore antigen specificity, remain unchanged.
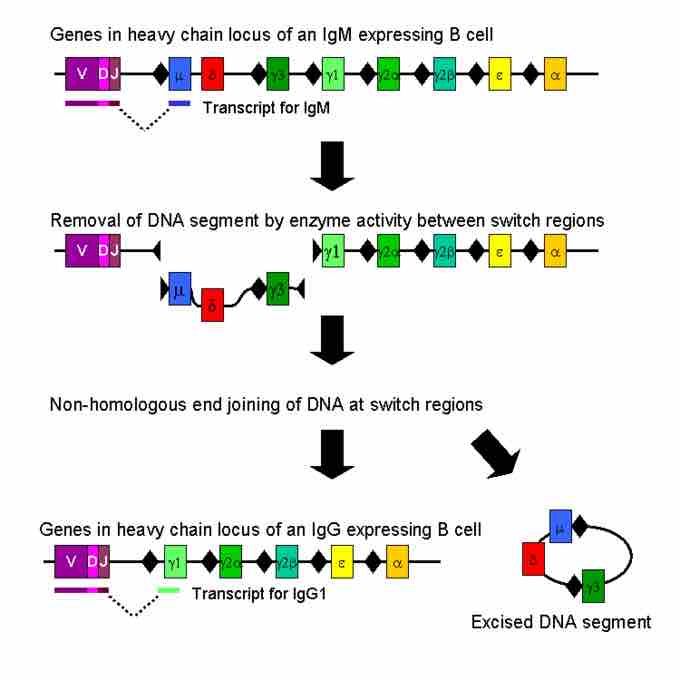
Class switch recombination
Mechanism of class switch recombination that allows isotype switching in activated B cells.
The progeny of a single B cell can produce antibodies, all specific for the same antigen, but with the ability to produce the effector function appropriate for each antigenic challenge. Class switching is triggered by cytokines; the isotype generated depends on which cytokines are present in the B cell environment. Class switching occurs in the heavy chain gene locus by a mechanism called class switch recombination (CSR). This mechanism relies on conserved nucleotide motifs, called switch (S) regions, found in DNA upstream of each constant region gene (except in the δ-chain). The DNA strand is broken by the activity of a series of enzymes at two selected S-regions. The variable domain exon is rejoined through a process called non-homologous end joining (NHEJ) to the desired constant region (γ, α or ε). This process results in an immunoglobulin gene that encodes an antibody of a different isotype.
Affinity Designations
A group of antibodies can be called monovalent (or specific) if they have affinity for the same epitope, or for the same antigen (but potentially different epitopes on the molecule), or for the same strain of microorganism (but potentially different antigens on or in it). In contrast, a group of antibodies can be called polyvalent (or unspecific) if they have affinity for various antigens or microorganisms. Intravenous immunoglobulin, if not otherwise noted, consists of polyvalent IgG. In contrast, monoclonal antibodies are monovalent for the same epitope.