The Protein Synthesis Machinery
In addition to the mRNA template, many molecules and macromolecules contribute to the process of translation. The composition of each component may vary across species. For instance, ribosomes may consist of different numbers of rRNAs and polypeptides depending on the organism. However, the general structures and functions of the protein synthesis machinery are comparable from bacteria to archaea to human cells. Translation requires the input of an mRNA template, ribosomes, tRNAs, and various enzymatic factors.
Ribosomes
A ribosome is a complex macromolecule composed of structural and catalytic rRNAs, and many distinct polypeptides. In eukaryotes, the synthesis and assembly of rRNAs occurs in the nucleolus.
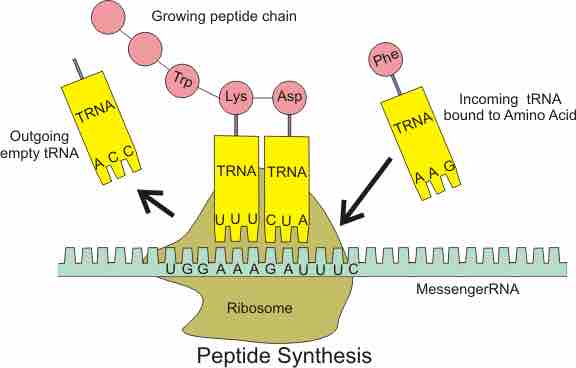
The ribosome in action
Structure and role of ribosomes during translation
Ribosomes exist in the cytoplasm in prokaryotes and in the cytoplasm and on rough endoplasmic reticulum membranes in eukaryotes. Mitochondria and chloroplasts also have their own ribosomes, and these look more similar to prokaryotic ribosomes (and have similar drug sensitivities) than the cytoplasmic ribosomes. Ribosomes dissociate into large and small subunits when they are not synthesizing proteins and reassociate during the initiation of translation.E. coli have a 30S small subunit and a 50S large subunit, for a total of 70S when assembled (recall that Svedberg units are not additive). Mammalian ribosomes have a small 40S subunit and a large 60S subunit, for a total of 80S. The small subunit is responsible for binding the mRNA template, whereas the large subunit sequentially binds tRNAs.
In bacteria, archaea, and eukaryotes, the intact ribosome has three binding sites that accomodate tRNAs: The A site, the P site, and the E site. Incoming aminoacy-tRNAs (a tRNA with an amino acid covalently attached is called an aminoacyl-tRNA) enter the ribosome at the A site. The peptidyl-tRNA carrying the growing polypeptide chain is held in the P site. The E site holds empty tRNAs just before they exit the ribosome.

Ribosome structure
The large ribosomal subunit sits atop the small ribosomal subunit and the mRNA is threaded through a groove near the interface of the two subunits. The intact ribosome has three tRNA binding sites: the A site for incoming aminoacyl-tRNAs; the P site for the peptidyl-tRNA carrying the growing polypeptide chain; and the E site where empty tRNAs exit (not shown in this figure but immediately adjacent to the P site.)
Each mRNA molecule is simultaneously translated by many ribosomes, all reading the mRNA from 5' to 3' and synthesizing the polypeptide from the N terminus to the C terminus. The complete mRNA/poly-ribosome structure is called a polysome.
tRNAs in eukaryotes
The tRNA molecules are transcribed by RNA polymerase III. Depending on the species, 40 to 60 types of tRNAs exist in the cytoplasm. Specific tRNAs bind to codons on the mRNA template and add the corresponding amino acid to the polypeptide chain. (More accurately, the growing polypeptide chain is added to each new amino acid bound in by a tRNA.)
The transfer RNAs (tRNAs) are structural RNA molecules. In eukaryotes, tRNA mole are transcribed from tRNA genes by RNA polymerase III. Depending on the species, 40 to 60 types of tRNAs exist in the cytoplasm. Serving as adaptors, specific tRNAs bind to sequences on the mRNA template and add the corresponding amino acid to the polypeptide chain. (More accurately, the growing polypeptide chain is added to each new amino acid brought in by a tRNA.) Therefore, tRNAs are the molecules that actually "translate" the language of RNA into the language of proteins.
Of the 64 possible mRNA codons (triplet combinations of A, U, G, and C) three specify the termination of protein synthesis and 61 specify the addition of amino acids to the polypeptide chain. Of the three termination codons, one (UGA) can also be used to encode the 21st amino acid, selenocysteine, but only if the mRNA contains a specific sequence of nucleotides known as a SECIS sequence. Of the 61 non-termination codons, one codon (AUG) also encodes the initiation of translation.
Each tRNA polynucleotide chain folds up so that some internal sections basepair with other internal sections. If just diagrammed in two dimensions, the regions where basepairing occurs are called stems, and the regions where no basepairs form are called loops, and the entire pattern of stems and loops that forms for a tRNA is called the "cloverleaf" structure. All tRNAs fold into very similar cloverleaf structures of four major stems and three major loops.

The two-dimensional cloverleaf structure of a typical tRNA.
All tRNAs, regardless of the species they come from or the amino acid they carry, self-basepair to produce a cloverleaf structure of four main stems and three main loops. The amino acid carried by the tRNA is covalently attached to the nucleotide at the 3' end of the tRNA, known as the tRNA's acceptor arm. The opposite end of the folded tRNA has the anticodon loop where the tRNA will basepair to the mRNA codon.
If viewed as a three-dimensional structure, all the basepaired regions of the tRNA are helical, and the tRNA folds into a L-shaped structure.
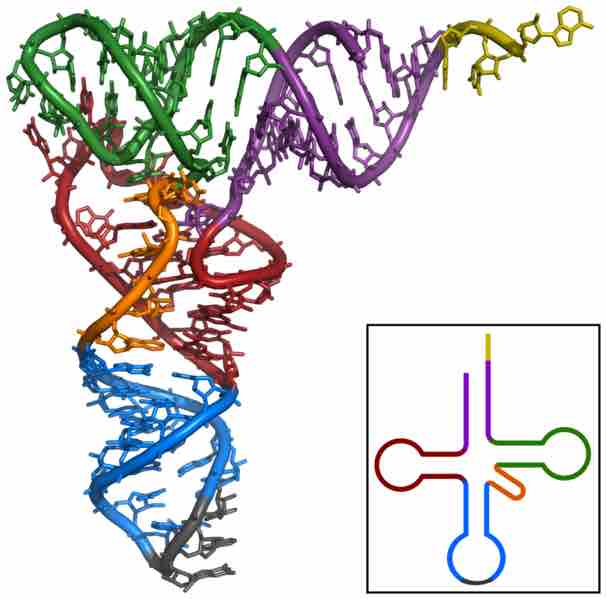
The three dimensional shape taken by tRNAs.
If viewed as a three-dimensional structure, all tRNAs are partially helical molecules that are vaguely L-shaped. The anticodon-containing loop is at one end of the molecule (in grey here) and the amino acid acceptor arm is at the other end of the molecule (in yellow here) past the bend of the "L".
Each tRNA has a sequence of three nucleotides located in a loop at one end of the molecule that can basepair with an mRNA codon. This is called the tRNA's anticodon. Each different tRNA has a different anticodon. When the tRNA anticodon basepairs with one of the mRNA codons, the tRNA will add an amino acid to a growing polypeptide chain or terminate translation, according to the genetic code. For instance, if the sequence CUA occurred on a mRNA template in the proper reading frame, it would bind a tRNA with an anticodon expressing the complementary sequence, GAU. The tRNA with this anticodon would be linked to the amino acid leucine.
Aminoacyl tRNA Synthetases
The process of pre-tRNA synthesis by RNA polymerase III only creates the RNA portion of the adaptor molecule. The corresponding amino acid must be added later, once the tRNA is processed and exported to the cytoplasm. Through the process of tRNA "charging," each tRNA molecule is linked to its correct amino acid by a group of enzymes called aminoacyl tRNA synthetases. When an amino acid is covalently linked to a tRNA, the resulting complex is known as an aminoacyl-tRNA. At least one type of aminoacyl tRNA synthetase exists for each of the 21 amino acids; the exact number of aminoacyl tRNA synthetases varies by species. These enzymes first bind and hydrolyze ATP to catalyze the formation of a covalent bond between an amino acid and adenosine monophosphate (AMP); a pyrophosphate molecule is expelled in this reaction. This is called "activating" the amino acid. The same enzyme then catalyzes the attachment of the activated amino acid to the tRNA and the simultaneous release of AMP. After the correct amino acid covalently attached to the tRNA, it is released by the enzyme. The tRNA is said to be charged with its cognate amino acid. (the amino acid specified by its anticodon is a tRNA's cognate amino acid.)