Purines are biologically synthesized as nucleotides and in particular as ribotides; i.e., bases attached to ribose 5-phosphate. A key regulatory step is the production of 5-phospho-α-D-ribosyl 1-pyrophosphate (PRPP) by PRPP synthetase, which is activated by inorganic phosphate and inactivated by purine ribonucleotides. It is not a committed step to purine synthesis because PRPP is also used in pyrimidine synthesis and salvage pathways. The first committed step is the reaction of PRPP, glutamine and water to 5'-phosphoribosylamine, glutamate, and pyrophosphate - catalyzed by pyrophosphate amidotransferase, which is activated by PRPP and inhibited by AMP, GMP and IMP.
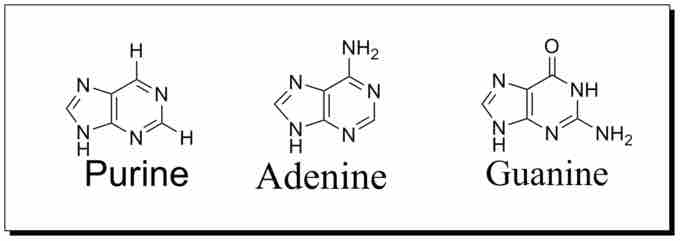
Purine structure
A purine is a nucleotide (a nucleoside + phosphate group) that is amine based and planar, aromatic, and heterocyclic. The structure of purine is that of a cyclohexane(pyrimidine group) and cyclopentane(imidazole group) attached to one another; the Nitrogen atoms are at positions 1,3,7,9. Adenine(A) and Guanine(G) are examples of purines which are involved in the construction of the backbone of the DNA and RNA.
Both adenine and guanine are derived from the nucleotide inosine monophosphate (IMP), which is the first compound in the pathway to have a completely formed purine ring system. Inosine monophosphate is synthesized on a pre-existing ribose-phosphate through a complex pathway. The carbon and nitrogen atoms of the purine ring, 5 and 4 respectively, come from multiple sources. The amino acid glycine contributes all its carbon (2) and nitrogen (1) atoms, with additional nitrogen atoms coming from glutamine (2) and aspartic acid (1), and additional carbon atoms coming from formyl groups (2). These are transferred from the coenzyme tetrahydrofolate as 10-formyltetrahydrofolate, along with a carbon atom from bicarbonate (1). Formyl groups build carbon-2 and carbon-8 in the purine ring system, which are the ones acting as bridges between two nitrogen atoms.
Unlike purines, pyrimidines are assembled before being attached to 5-phosphoribosyl-1-pyrophosphate (PRPP). The first regulated step in pyrimidine biosynthesis is the formation of carbamoyl phosphate by carbamoyl phosphate synthetase II. This is then converted by carbamoyl aspartic acid by aspartic transcarbamolyase (aspartate carbamoyl transferase), which will be dehydrated to dihydroorotate by dihydroorotase. Dihydroorotate then enters the mitochondria, where it is oxidised into orotate through removal of hydrogens by dihydroorotate dehydrogenase. This is the only mitochondrial step in nucleotide rings biosynthesis. Orotate is then converted to OMP by orotate phosphoribosyltransferase. OMP will then be decarboxylated to UMP by OMP decarboxylase. UMP in turn will be converted to UDP by phosphorylation via uridine-cytidine kinase 2. UDP in turn will be phosphorylated to UTP by nucleoside diphosphate kinaseuridine 5'-triphosphate(UTP). UDP can also be converted to CTP by CTP synthase cytidine 5'triphosphate (CTP)using glutamine and ATP.The first three enzymes are all coded by the same gene in Metazoa (CAD).
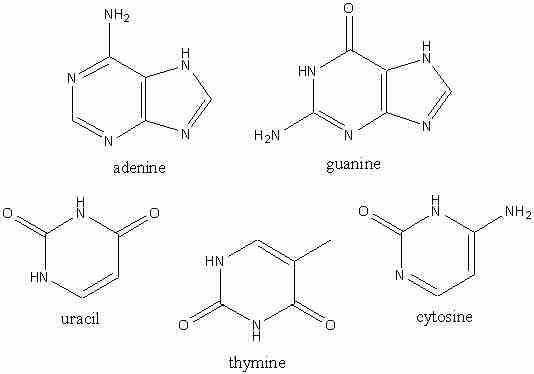
What forms a nucleic acid structure?
The four nitrogenous bases present in DNA are adenine (A), guanine (G), cytosine (C) and thymine (T). Adenine and guanine are purines and cytosine and thymine are pyrimidines.
In Fungi, a similar protein exists but lacks the dihydroorotase function: another protein catalyzes the second step. In other organisms (Bacteria, Archaea and the other Eukaryota), the first three steps are done by three different enzymes. CTP synthase (or CTP synthetase) is an enzyme involved in pyrimidine biosynthesis. It intraconverts UTP and CTP. The source of the amine/amino group in CTP is glutamine. CTP synthase is activated by GTP, a purine. This acts to balance the relative amounts of purine and pyrimidine nucleotides. CTP synthase is inhibited by reversible by CTP and irreversible for example by the glutamine analogon DON. The following human genes encode proteins that possess CTP synthase activity: