As a genus, Chloroflexus spp. are Gram-negative filamentous anoxygenic phototrophic (FAP) organisms that utilize type II photosynthetic reaction centers containing bacteriochlorophyll a similar to the purple bacteria, and light-harvesting chlorosomes containing bacteriochlorophyll c similar to green sulfur bacteria of the Chlorobi. As the name implies, these anoxygenic phototrophs do not produce oxygen as a byproduct of photosynthesis, in contrast to oxygenic phototrophs such as cyanobacteria, algae, and higher plants. While oxygenic phototrophs use water as an electron donor for phototrophy, Chloroflexus uses reduced sulfur compounds such as hydrogen sulfide, thiosulfate, or elemental sulfur. This belies their antiquated name green non-sulfur bacteria. However, Chloroflexus spp. can also utilize hydrogen (H2) as a source of electrons.
Chloroflexus aurantiacus is thought to grow photoheterotrophically in nature, but it has the capability of fixing inorganic carbon through photoautotrophic growth. Instead of using the Calvin-Benson-Bassham Cycle typical of plants, Chloroflexus aurantiacus has been demonstrated to use a novel autotrophic pathway known as the 3-Hydroxypropionate pathway .The complete electron transport chain for Chloroflexus spp. is not yet known. Particularly, Chloroflexus aurantiacus has not been demonstrated to have a cytochrome bc1 complex. It may use different proteins to reduce cytochrome c.
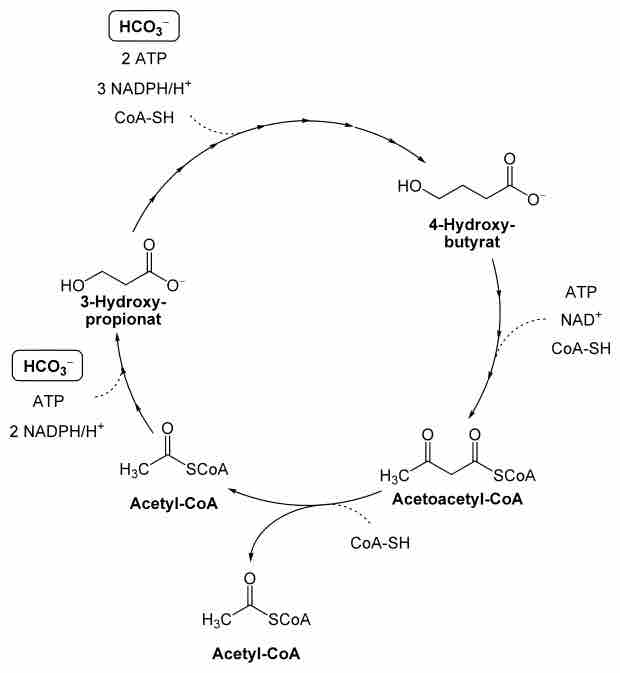
3-Hydroxypropionate Pathway
A schematic representation of the 3-Hydroxypropionate CO2 assimilatory pathway observed in Chloroflexus bacteria.
Chloroflexus aurantiacus is a photosynthetic bacterium isolated from hot springs, belonging to the green non-sulfur bacteria. This organism is thermophilic and can grow at temperatures from 35 °C to 70 °C. Chloroflexus aurantiacus can survive in the dark if oxygen is available. When grown in the dark, Chloroflexus aurantiacus has a dark orange color. When grown in sunlight it is dark green. The individual bacteria tend to form filamentous colonies enclosed in sheaths, which are known as trichomes. One of the main reasons for interest in Chloroflexus aurantiacus is in the study of the evolution of photosynthesis.
How did photosynthesis arise in bacteria? The answer to this question is complicated by the fact that there are several types of light-harvesting energy capture systems. Chloroflexus aurantiacus has been of interest in the search for origins of the so-called type II photosynthetic reaction center. One idea is that bacteria with respiratory electron transport evolved photosynthesis by coupling a light-harvesting energy capture system to the pre-existing respiratory electron transport chain. Therefore, rare organisms like Chloroflexus aurantiacus that can survive using either respiration or photosynthesis are of interest in on-going attempts to trace the evolution of photosynthesis.
The Chloroflexi or Chlorobacteria are a phylum of bacteria containing isolates with a diversity of phenotypes including members that are aerobic thermophiles, which use oxygen and grow well in high temperatures, anoxygenic phototrophs, which use light for photosynthesis, and anaerobic halorespirers, which use halogenated organics (such as the toxic chlorinated ethenes and polychlorinated biphenyls) as energy sources. Whereas most bacteria, in terms of diversity, are diderms and stain Gram negative with the exception of the Firmicutes (low CG Gram positives), Actinobacteria (high CG gram positives), and the Deinococcus-Thermus group (Gram positive, but diderms with thick peptidoglycan), the members of the phylum Chloroflexi are monoderms and stain mostly Gram negative.
In 1987, Carl Woese, regarded as the forerunner of the molecular phylogeny revolution, divided Eubacteria into 11 divisions based on 16S ribosomal RNA (SSU) sequences and grouped the genera Chloroflexus, Herpetosiphon, and Thermomicrobium into the "Green non-sulfur bacteria and relatives," which was temporarily renamed as "Chloroflexi" in Volume One of Bergey's Manual of Systematic Bacteriology.
Recent phylogenetic analysis of the Chloroflexi has found very weak support for the grouping together of the different classes currently part of the phylum. The six classes that make up the phylum did not consistently form a well-supported monophyletic clade in phylogenetic trees based on concatenated sequences for large datasets of proteins. No conserved signature indels were identified that were uniquely shared by the entire phylum. However, the classes "Chloroflexi" and Thermomicrobia were found to group together consistently by both phylogenetic means and the identification of shared conserved signature indels in the 50S ribosomal protein L19 and the enzyme UDP-glucose 4-epimerase. It has been suggested that the phylum Chloroflexi "sensu stricto" should comprise only the classes Chloroflexi and Thermomicrobia, and the other four classes ("Dehalococcoidetes," Anaerolineae, Caldilineae, and Ktedonobacteria) may represent one or more independent phyla branching in the neighborhood of the Chloroflexi.