Variations in Size and Number of Genes
Genetic diversity refers to any variation in the nucleotides, genes, chromosomes, or whole genomes of organisms. Genetic diversity at its most elementary level is represented by differences in the sequences of nucleotides (adenine, cytosine, guanine, and thymine) that form the DNA (deoxyribonucleic acid) within the cells of the organism. The DNA is contained in the chromosomes present within the cell; some chromosomes are contained within specific organelles in the cell (for example, the chromosomes of mitochondria and chloroplast). Nucleotide variation is measured for discrete sections of the chromosomes, called genes. Thus, each gene compromises a hereditary section of DNA that occupies a specific place of the chromosome, and controls a particular characteristic of an organism.
Chromosomes
Most organisms are diploid, having two sets of chromosomes, and therefore two copies (called alleles) of each gene. However, some organisms can be haploid, triploid, or tetraploid (having one, three, or four sets of chromosomes respectively). Within any single organism, there may be variation between the two (or more) alleles for each gene. This variation is introduced either through mutation of one of the alleles, or as a result of sexual reproduction.
During sexual reproduction, offspring inherit alleles from both parents and these alleles might be slightly different, especially if there has been migration or hybridization of organisms, so that the parents may come from different populations and gene pools. Also, when the offspring's chromosomes are copied after fertilization, genes can be exchanged in a process called sexual recombination. Harmless mutations and sexual recombination may allow the evolution of new characteristics.
Genome Size and Number
Genome size is usually measured in base pairs (or bases in single-stranded DNA or RNA). The C-value is another measure of genome size. The C-value refers to the amount, in picograms, of DNA contained within a haploid nucleus (e.g. a gamete) or one half the amount in a diploid somatic cell of a eukaryotic organism. In some cases (notably among diploid organisms), the terms C-value and genome size are used interchangeably, however in polyploids the C-value may represent two or more genomes contained within the same nucleus.
Different species can have different numbers of genes within the entire DNA or genome of the organism. However, a greater total number of genes might not correspond with a greater observable complexity in the anatomy and physiology of the organism (i.e. greater phenotypic complexity). For example, the predicted size of the human genome is not much larger than the genomes of some invertebrates and plants, and may even be smaller than the Indian rice genome. In humans, more proteins are encoded per gene than in other species. In prokaryotic genomes, research has shown that there is a significant positive correlation between the C-value of prokaryotes and the amount of genes that compose the genome. This indicates that gene number is the main factor influencing the size of the prokaryotic genome.
Genes vs Genome Size
In eukaryotic organisms, there is a paradox observed, namely that the number of genes that make up the genome does not correlate with genome size. In other words, the genome size is much larger than would be expected given the total number of protein coding genes . Genome size can increase by duplication, insertion, or polyploidization and the process of recombination can lead to both DNA loss or gain. It is also possible that genomes can shrink due to deletions.
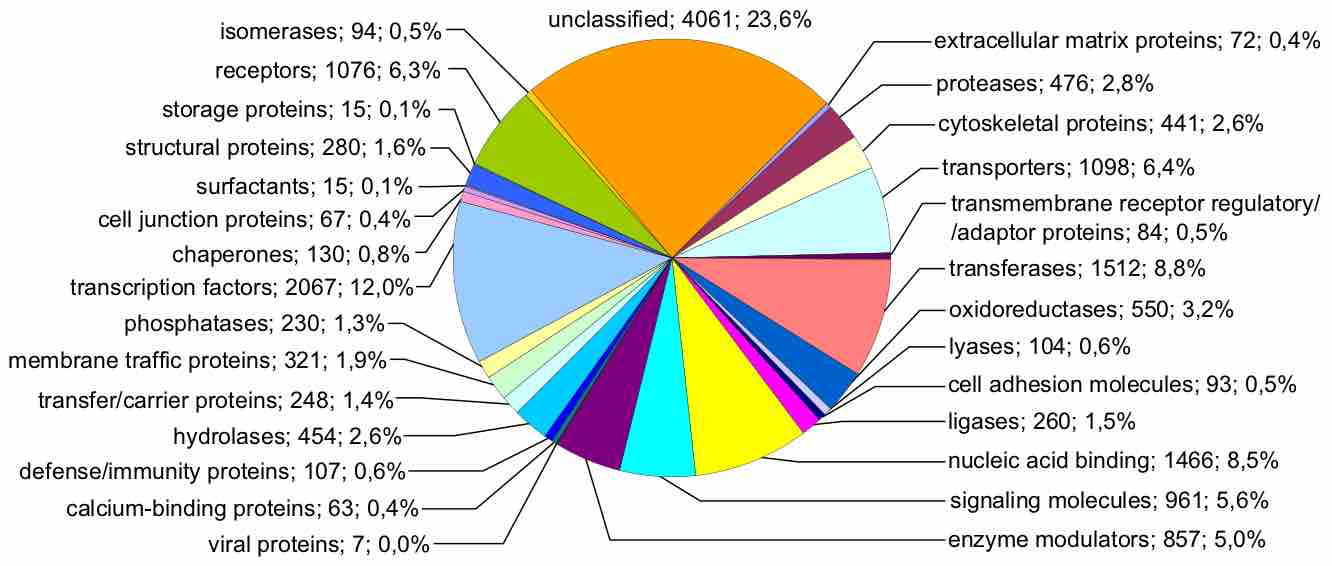
Gene variation in the Genome
This figure represents the human genome, categorized by function of each gene product, given both as number of genes and as percentage of all genes. Importantly, genome size does not necessarily correlate with complexity.
A famous example for such gene decay is the genome of Mycobacterium leprae, the causative agent of leprosy. M.leprae has lost many once-functional genes over time due to the formation of pseudogenes. This is evident in looking at its closest ancestor Mycobacterium tuberculosis. M. leprae lives inside and replicates inside of a host and due to this arrangement it does not have a need for many of the genes it once carried which allowed it to live and prosper outside of the host. Thus over time these genes have lost their function through mechanisms such as mutation causing them to become pseudogenes. It is beneficial to an organism to rid itself of non-essential genes because it makes replicating its DNA much faster and more energy-efficient.
An example of increasing genome size over time is seen in filamentous plant pathogens. These plant pathogen genomes have been growing larger over the years due to repeat-driven expansion. The repeat-rich regions contain genes coding for host interaction proteins. With the addition of more and more repeats to these regions the plants increase the possibility of developing new virulence factors through mutation and other forms of genetic recombination. In this way it is beneficial for these plant pathogens to have larger genomes.