Accumulating Changes Over Time
The evolution of the genome is characterized by the accumulation of changes. The analaysis of genomes and their changes in sequence or size over time involves various fields. There are various mechanisms that have contributed to genome evolution and these include gene and genome duplications, polyploidy, mutation rates, transposable elements, pseudogenes, exon shuffling and genomic reduction and gene loss. The concepts of gene and whole-genome duplication are discussed as their own independent concepts, thus, the focus will be on other mechanisms.
Mutation Rates
Mutation rates differ between species and even between different regions of the genome of a single species . Spontaneous mutations often occur which can cause various changes in the genome. Mutations can result in the addition or deletion of one or more nucleotide bases. A change in the code can result in a frameshift mutation which causes the entire code to be read in the wrong order and thus often results in a protein becoming non-functional. A mutation in a promoter region, enhancer region or a region coding for transcription factors can also result in either a loss of function or and upregulation or downregulation in transcription of that gene. Mutations are constantly occurring in an organism's genome and can cause either a negative effect, positive effect or no effect at all.
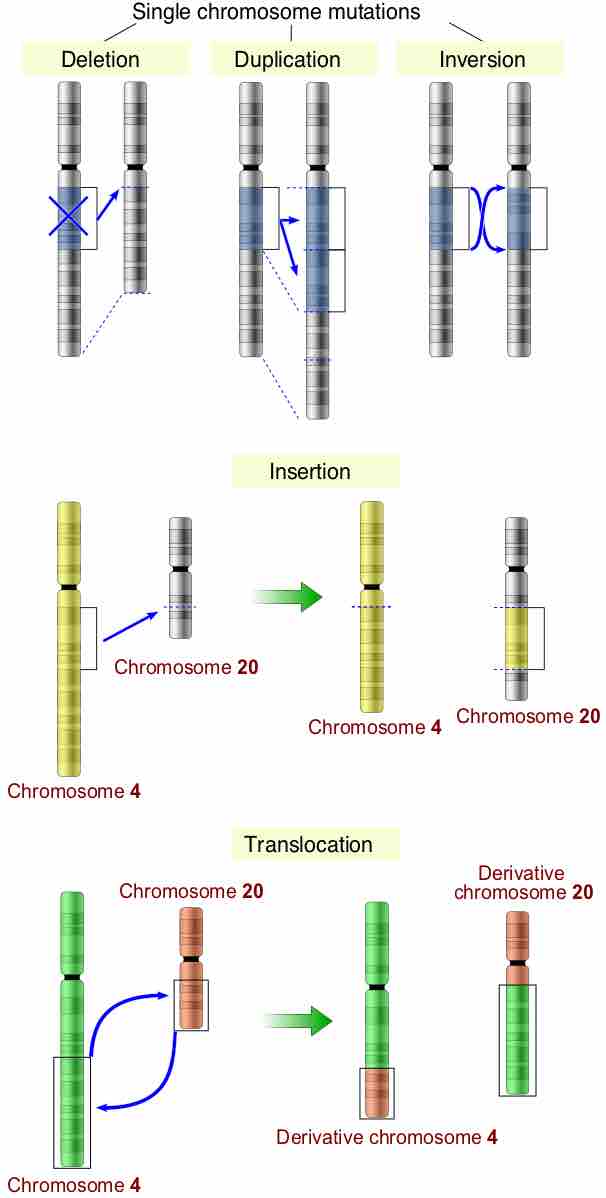
Chromosomal Mutations
Chromosomal mutations over time can accumulate and promote diversity and evolution if a produced trait is favorable.
Transposable Elements
Transposable elements are regions of DNA that can be inserted into the genetic code through one of two mechanisms. These mechanisms work similarly to "cut-and-paste" and "copy-and-paste" functionalities in word processing programs. The "cut-and-paste" mechanism works by excising DNA from one place in the genome and inserting itself into another location in the code. The "copy-and-paste" mechanism works by making a genetic copy or copies of a specific region of DNA and inserting these copies elsewhere in the code. The most common transposable element in the human genome is the Alu sequence, which is present in the genome over one million times.
Pseudogenes
Often a result of spontaneous mutation, pseudogenes are dysfunctional genes derived from previously functional gene relatives. There are many mechanisms by which a functional gene can become a pseudogene including the deletion or insertion of one or multiple nucleotides. This can result in a shift of reading frame, causing the gene to longer code for the expected protein, a premature stop codon or a mutation in the promoter region. Often cited examples of pseudogenes within the human genome include the once functional olfactory gene families. Over time, many olfactory genes in the human genome became pseudogenes and were no longer able to produce functional proteins, explaining the poor sense of smell humans possess in comparison to their mammalian relatives.
Exon Shuffling
Exon shuffling is a mechanism by which new genes are created. This can occur when two or more exons from different genes are combined together or when exons are duplicated. Exon shuffling results in new genes by altering the current intron-exon structure. This can occur by any of the following processes: transposon mediated shuffling, sexual recombination or illegitimate recombination. Exon shuffling may introduce new genes into the genome that can be either selected against and deleted or selectively favored and conserved.
Genome Reduction and Gene Loss
Many species exhibit genome reduction when subsets of their genes are not needed anymore. This typically happens when organisms adapt to a parasitic life style, e.g. when their nutrients are supplied by a host. As a consequence, they lose the genes need to produce these nutrients. In many cases, there are both free living and parasitic species that can be compared and their lost genes identified. Good examples are the genomes of Mycobacterium tuberculosis and Mycobacterium leprae, the latter of which has a dramatically reduced genome. Another beautiful example are endosymbiont species. For instance, Polynucleobacter necessarius was first described as a cytoplasmic endosymbiont of the ciliate Euplotes aediculatus. The latter species dies soon after being cured of the endosymbiont. In the few cases in which P. necessarius is not present, a different and rarer bacterium apparently supplies the same function. No attempt to grow symbiotic P. necessarius outside their hosts has yet been successful, strongly suggesting that the relationship is obligate for both partners. Yet, closely related free-living relatives of P. necessarius have been identified. The endosymbionts have a significantly reduced genome when compared to their free-living relatives (1.56 Mbp vs. 2.16 Mbp).