Genetic Drift vs. Natural Selection
Genetic drift is the converse of natural selection. The theory of natural selection maintains that some individuals in a population have traits that enable to survive and produce more offspring, while other individuals have traits that are detrimental and may cause them to die before reproducing. Over successive generation, these selection pressures can change the gene pool and the traits within the population. For example, a big, powerful male gorilla will mate with more females than a small, weak male and therefore more of his genes will be passed on to the next generation. His offspring may continue to dominate the troop and pass on their genes as well. Over time, the selection pressure will cause the allele frequencies in the gorilla population to shift toward large, strong males.
Unlike natural selection, genetic drift describes the effect of chance on populations in the absence of positive or negative selection pressure. Through random sampling, or the survival or and reproduction of a random sample of individuals within a population, allele frequencies within a population may change. Rather than a male gorilla producing more offspring because he is stronger, he may be the only male available when a female is ready to mate. His genes are passed on to future generation because of chance, not because he was the biggest or the strongest. Genetic drift is the shift of alleles within a population due to chance events that cause random samples of the population to reproduce or not.
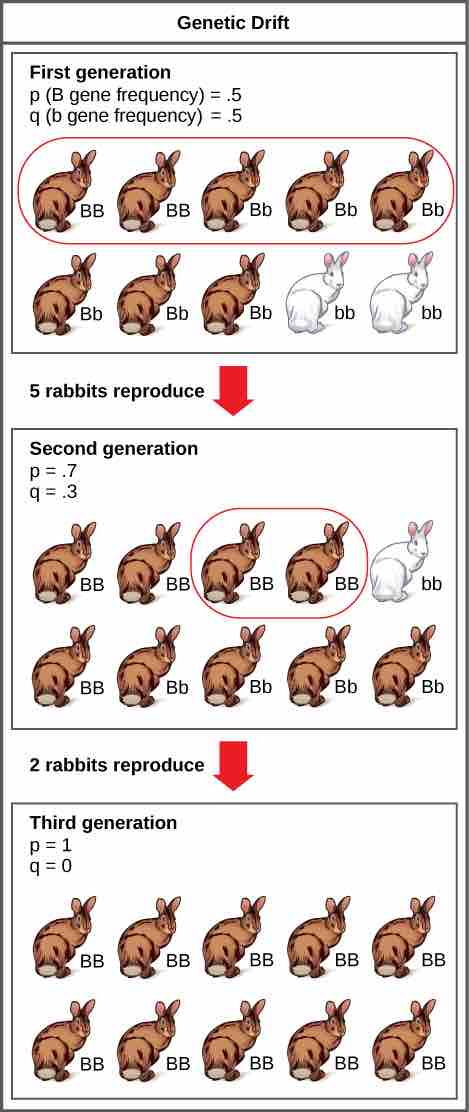
Effect of genetic drift
Genetic drift in a population can lead to the elimination of an allele from that population by chance. In this example, the brown coat color allele (B) is dominant over the white coat color allele (b). In the first generation, the two alleles occur with equal frequency in the population, resulting in p and q values of .5. Only half of the individuals reproduce, resulting in a second generation with p and q values of .7 and .3, respectively. Only two individuals in the second generation reproduce and, by chance, these individuals are homozygous dominant for brown coat color. As a result, in the third generation the recessive b allele is lost.
Small populations are more susceptible to the forces of genetic drift. Large populations, on the other hand, are buffered against the effects of chance. If one individual of a population of 10 individuals happens to die at a young age before leaving any offspring to the next generation, all of its genes (1/10 of the population's gene pool) will be suddenly lost. In a population of 100, that individual represents only 1 percent of the overall gene pool; therefore, genetic drift has much less impact on the larger population's genetic structure.
The Bottleneck Effect
Genetic drift can also be magnified by natural events, such as a natural disaster that kills a large portion of the population at random. The bottleneck effect occurs when only a few individuals survive and reduces variation in the gene pool of a population. The genetic structure of the survivors becomes the genetic structure of the entire population, which may be very different from the pre-disaster population.
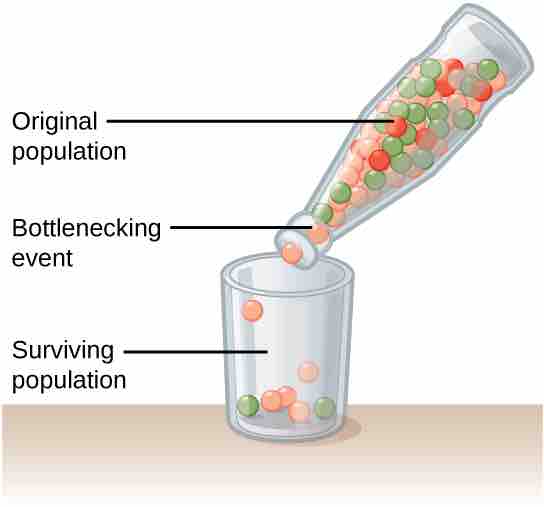
Effect of a bottleneck on a population
A chance event or catastrophe can reduce the genetic variability within a population.
The Founder Effect
Another scenario in which populations might experience a strong influence of genetic drift is if some portion of the population leaves to start a new population in a new location or if a population gets divided by a physical barrier of some kind. In this situation, it is improbable that those individuals are representative of the entire population, which results in the founder effect. The founder effect occurs when the genetic structure changes to match that of the new population's founding fathers and mothers.
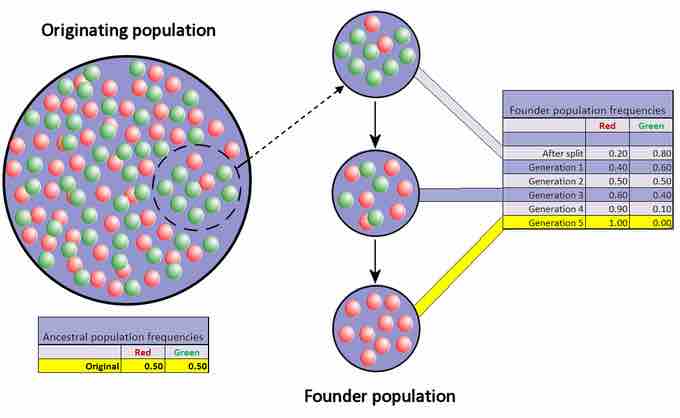
The Founder Effect
The founder effect occurs when a portion of the population (i.e. "founders") separates from the old population to start a new population with different allele frequencies.
The founder effect is believed to have been a key factor in the genetic history of the Afrikaner population of Dutch settlers in South Africa, as evidenced by mutations that are common in Afrikaners, but rare in most other populations. This was probably due to the fact that a higher-than-normal proportion of the founding colonists carried these mutations. As a result, the population expresses unusually high incidences of Huntington's disease (HD) and Fanconi anemia (FA), a genetic disorder known to cause blood marrow and congenital abnormalities, even cancer.
Drift and fixation
The Hardy–Weinberg principle states that within sufficiently large populations, the allele frequencies remain constant from one generation to the next unless the equilibrium is disturbed by migration, genetic mutation, or selection.
Because the random sampling can remove, but not replace, an allele, and because random declines or increases in allele frequency influence expected allele distributions for the next generation, genetic drift drives a population towards genetic uniformity over time. When an allele reaches a frequency of 1 (100%) it is said to be "fixed" in the population and when an allele reaches a frequency of 0 (0%) it is lost. Once an allele becomes fixed, genetic drift for that allele comes to a halt, and the allele frequency cannot change unless a new allele is introduced in the population via mutation or gene flow. Thus even while genetic drift is a random, directionless process, it acts to eliminate genetic variation over time.
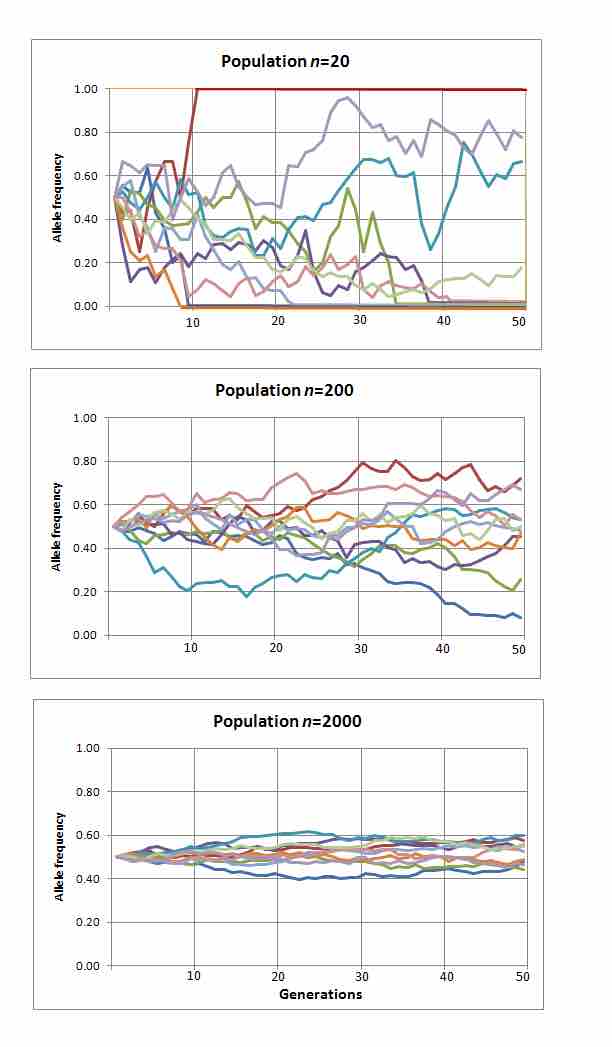
Genetic drift over time
Ten simulations of random genetic drift of a single given allele with an initial frequency distribution 0.5 measured over the course of 50 generations, repeated in three reproductively synchronous populations of different sizes. In these simulations, alleles drift to loss or fixation (frequency of 0.0 or 1.0) only in the smallest population.Effect of population size on genetic drift: Ten simulations each of random change in the frequency distribution of a single hypothetical allele over 50 generations for different sized populations; first population size n=20, second population n=200, and third population n=2000.