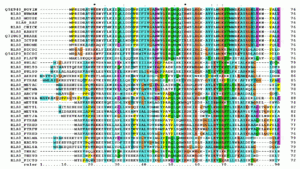
Multiple sequence alignment
Background to the schools Wikipedia
SOS Children offer a complete download of this selection for schools for use on schools intranets. SOS Child sponsorship is cool!
A multiple sequence alignment (MSA) is a sequence alignment of three or more biological sequences, generally protein, DNA, or RNA. In general, the input set of query sequences are assumed to have an evolutionary relationship by which they share a lineage and are descended from a common ancestor. From the resulting MSA, sequence homology can be inferred and phylogenetic analysis can be conducted to assess the sequences' shared evolutionary origins. Visual depictions of the alignment as in the image at right illustrate mutation events such as point mutations (single amino acid or nucleotide changes) that appear as differing characters in a single alignment column, and insertion or deletion mutations (or indels) that appear as gaps in one or more of the sequences in the alignment. Multiple sequence alignment is often used to assess sequence conservation of protein domains, tertiary and secondary structures, and even individual amino acids or nucleotides.
Multiple sequence alignment also refers to the process of aligning such a sequence set. Because three or more sequences of biologically relevant length can be difficult and are almost always time-consuming to align by hand, computational algorithms are used to produce and analyze the alignments. MSAs require more sophisticated methodologies than pairwise alignment because they are more computationally complex to produce. Most multiple sequence alignment programs use heuristic methods rather than global optimization because identifying the optimal alignment between more than a few sequences of moderate length is prohibitively computationally expensive.
Dynamic programming and computational complexity
The most direct method for producing an MSA uses the dynamic programming technique to identify the globally optimal alignment solution. For proteins, this method usually involves two sets of parameters: a gap penalty and a substitution matrix assigning scores or probabilities to the alignment of each possible pair of amino acids based on the similarity of the amino acids' chemical properties and the evolutionary probability of the mutation. For nucleotide sequences a substitution matrix can be used, but since there are only four possible standard characters per sequence and the individual nucleotides do not typically differ much in substitution probability, the parameters for DNA and RNA sequences usually consist of a gap penalty, a positive score for character matches, and a negative score for mismatches.
For n individual sequences, the method requires constructing the n-dimensional equivalent of the matrix formed in standard pairwise dynamic programming. The search space thus increases exponentially with increasing n and is also strongly dependent on sequence length. To find the global optimum for n sequences this way has been shown to be an NP-complete problem. Methods to reduce the search space by first performing pairwise dynamic programming on each pair of sequences in the query set and searching only the solution space near these results (effectively finding the intersection between local paths immediately surrounding each pairwise optimum solution) render the dynamic programming technique more efficient. The so-called "sum of pairs" method has been implemented in the software package MSA, but it is still impractical for many MSA applications that require the simultaneous alignment of dozens or even a few hundred sequences. Dynamic programming methods are now used only when an extremely high-quality alignment of a small number of sequences is needed, and as a benchmarking standard in evaluating new or refined heuristic techniques.
Progressive alignment construction
One method of performing a heuristic alignment search is the progressive technique (also known as the hierarchical or tree method) that builds up a final MSA by first performing a series of pairwise alignments on successively less closely related sequences. Such methods begin by aligning the two most closely related sequences first and then successively aligning the next most closely related sequence in the query set to the alignment produced in the previous step. The initial "most related" pair is determined by an efficient clustering method such as neighbour-joining based on a simple heuristic search of the query set with a tool like FASTA. Progressive techniques therefore automatically construct a phylogenetic tree as well as an alignment.
One major limitation of progressive methods is their heavy dependence on the initial assignment of relatedness and on the quality of the initial alignment. The methods are thus sensitive as well to the distribution of sequences in the query set; performance improves when relatedness among query sequences is a relatively smooth gradient rather than distantly separated clusters. Performance also degrades significantly when all of the sequences in the set are rather distantly related, because inaccuracies in the initial alignment are then more likely. Most modern progressive methods modify their scoring function with a secondary weighting function that assigns scaling factors to individual members of the query set in a nonlinear fashion based on their phylogenetic distance from their nearest neighbors. Judicious choice of weighting can aid in evaluating relatedness and mitigate the effects of relatively poor initial alignments early in the progression.
Progressive alignment methods are efficient enough to implement on a large scale for many sequences and are often run on publicly accessible web servers so users need not locally install the applications of interest. A very popular progressive alignment method is the Clustal family, especially the weighted variant ClustalW to which access is provided by a large number of web portals including GenomeNet, EBI, and EMBNet. Different portals or implementations can vary in user interface and make different parameters accessible to the user. Clustal is used extensively for phylogenetic tree construction and as input for protein structure prediction by homology modeling.
Another common progressive alignment method called T-Coffee is slower than Clustal and its derivatives but generally produces more accurate alignments for distantly related sequence sets. T-Coffee calculates pairwise alignments by combining the direct alignment of the pair with indirect alignments that aligns each sequence of the pair to a third sequence. It uses the output from Clustal as well as another local alignment program LALIGN, which finds multiple regions of local alignment between two sequences. The resulting alignment and phylogenetic tree are used as a guide to produce new and more accurate weighting factors.
Because progressive methods are heuristics that are not guaranteed to converge to a global optimum, alignment quality can be difficult to evaluate and their true biological significance can be obscure. A very recent semi-progressive method that improves alignment quality and does not use a lossy heuristic while still running in polynomial time has been implemented in the program PSAlign.
Iterative methods
A set of methods to produce MSAs while reducing the errors inherent in progressive methods are classified as "iterative" because they work similarly to progressive methods but repeatedly realign the initial sequences as well as adding new sequences to the growing MSA. One reason progressive methods are so strongly dependent on a high-quality initial alignment is the fact that these alignments are always incorporated into the final result - that is, once a sequence has been aligned into the MSA, its alignment is not considered further. This approximation improves efficiency at the cost of accuracy. By contrast, iterative methods can return to previously calculated pairwise alignments or sub-MSAs incorporating subsets of the query sequence as a means of optimizing a general objective function such as finding a high-quality alignment score.
A variety of subtly different iteration methods have been implemented and made available in software packages; reviews and comparisons have been useful but generally refrain from choosing a "best" technique. The software package PRRN/PRRP uses a hill-climbing algorithm to optimize its MSA alignment score and iteratively corrects both alignment weights and locally divergent or "gappy" regions of the growing MSA. PRRP performs best when refining an alignment previously constructed by a faster method.
Another iterative program, DIALIGN, takes an unusual approach of focusing narrowly on local alignments between sub-segments or sequence motifs without introducing a gap penalty. The alignment of individual motifs is then achieved with a matrix representation similar to a dot-matrix plot in a pairwise alignment. An alternative method that uses fast local alignments as anchor points or "seeds" for a slower global-alignment procedure is implemented in the CHAOS/DIALIGN suite.
A third popular iteration-based method called MUSCLE (multiple sequence alignment by log-expectation) improves on progressive methods with a more accurate distance measure to assess the relatedness of two sequences. The distance measure is updated between iteration stages (although, in its original form, MUSCLE contained only 2-3 iterations depending on whether refinement was enabled).
Hidden Markov models
Hidden Markov models are probabilistic models that can assign likelihoods to all possible combinations of gaps, matches, and mismatches to determine the most likely MSA or set of possible MSAs. HMMs can produce a single highest-scoring output but can also generate a family of possible alignments that can then be evaluated for biological significance. Because HMMs are probabilistic, they do not produce the same solution every time they are run on the same dataset; thus they cannot be guaranteed to converge to an optimal alignment. HMMs can produce both global and local alignments. Although HMM-based methods have been developed relatively recently, they offer significant improvements in computational speed, especially for sequences that contain overlapping regions.
Typical HMM-based methods work by representing an MSA as a form of directed acyclic graph known as a partial-order graph, which consists of a series of nodes representing possible entries in the columns of an MSA. In this representation a column that is absolutely conserved (that is, that all the sequences in the MSA share a particular character at a particular position) is coded as a single node with as many outgoing connections as there are possible characters in the next column of the alignment. In the terms of a typical hidden Markov model, the observed states are the individual alignment columns and the "hidden" states represent the presumed ancestral sequence from which the sequences in the query set are hypothesized to have descended. An efficient search variant of the dynamic programming method, known as the Viterbi algorithm, is generally used to successively align the growing MSA to the next sequence in the query set to produce a new MSA. This is distinct from progressive alignment methods because the alignment of prior sequences is updated at each new sequence addition. However, like progressive methods, this technique can be influenced by the order in which the sequences in the query set are integrated into the alignment, especially when the sequences are distantly related.
Several software programs are available in which variants of HMM-based methods have been implemented and which are noted for their scalability and efficiency, although properly using an HMM method is more complex than using more common progressive methods. The simplest is POA (Partial-Order Alignment); a similar but more generalized method is implemented in the package SAM (Sequence Alignment and Modeling System). SAM has been used as a source of alignments for protein structure prediction to participate in the CASP structure prediction experiment and to develop a database of predicted proteins in the yeast species S. cerevisiae. HMM methods can also be used for database search with HMMER.
Genetic algorithms and simulated annealing
Standard optimization techniques in computer science - both of which were inspired by, but do not directly reproduce, physical processes - have also been used in an attempt to more efficiently produce quality MSAs. One such technique, genetic algorithms, has been used for MSA production in an attempt to broadly simulate the hypothesized evolutionary process that gave rise to the divergence in the query set. The method works by breaking a series of possible MSAs into fragments and repeatedly rearranging those fragments with the introduction of gaps at varying positions. A general objective function is optimized during the simulation, most generally the "sum of pairs" maximization function introduced in dynamic programming-based MSA methods. A technique for protein sequences has been implemented in the software program SAGA (Sequence Alignment by Genetic Algorithm) and its equivalent in RNA is called RAGA.
The technique of simulated annealing, by which an existing MSA produced by another method is refined by a series of rearrangements designed to find more optimal regions of alignment space than the one the input alignment already occupies. Like the genetic algorithm method, simulated annealing maximizes an objective function like the sum-of-pairs function. Simulated annealing uses a metaphorical "temperature factor" that determines the rate at which rearrangements proceed and the likelihood of each rearrangement; typical usage alternates periods of high rearrangement rates with relatively low likelihood (to explore more distant regions of alignment space) with periods of lower rates and higher likelihoods to more thoroughly explore local minima near the newly "colonized" regions. This approach has been implemented in the program MSASA (Multiple Sequence Alignment by Simulated Annealing).
Motif finding
Motif finding, also known as profile analysis, is a method of locating sequence motifs in global MSAs that is both a means of producing a better MSA and a means of producing a scoring matrix for use in searching other sequences for similar motifs. A variety of methods for isolating the motifs have been developed, but all are based on identifying short highly conserved patterns within the larger alignment and constructing a matrix similar to a substitution matrix that reflects the amino acid or nucleotide composition of each position in the putative motif. The alignment can then be refined using these matrices. In standard profile analysis, the matrix includes entries for each possible character as well as entries for gaps. Alternatively, statistical pattern-finding algorithms can identify motifs as a precursor to an MSA rather than as a derivation. In many cases when the query set contains only a small number of sequences or contains only highly related sequences, pseudocounts are added to normalize the distribution reflected in the scoring matrix. In particular, this corrects zero-probability entries in the matrix to values that are small but nonzero.
Blocks analysis is a method of motif finding that restricts motifs to ungapped regions in the alignment. Blocks can be generated from an MSA or they can be extracted from unaligned sequences using a precalculated set of common motifs previously generated from known gene families. Block scoring generally relies on the spacing of high-frequency characters rather than on the calculation of an explicit substitution matrix. The BLOCKS server provides an interactive method to locate such motifs in unaligned sequences.
Statistical pattern-matching has been implemented using both the expectation-maximization algorithm and the Gibbs sampler. One of the most common motif-finding tools, known as MEME, uses expectation maximization and hidden Markov methods to generate motifs that are then used as search tools by its companion MAST in the combined suite MEME/MAST.