The retroviral while in the viral capsid consists of a dimer RNA. It has a cap at the 5' end and a poly(A) tail at the 3' end. The RNA genome also has terminal noncoding regions, which are important in replication, and internal regions that encode virion proteins for gene expression.
The 5' end includes four regions, which are R, U5, PBS, and L.
- The R region is a short repeated sequence at each end of the genome during the reverse transcription in order to ensure correct end-to-end transfer in growing chain.
- U5, on the other hand, is a short unique sequence between R and PBS.
- PBS (primer binding site) consists of 18 bases complementary to 3' end of the tRNA primer, which supplies an 'OH group to initiate reverse transcription.
- The L region is an untranslated leader region that give the signal for packaging of genome RNA.
The 3' end includes 3 regions, which are PPT (polypurine tract), U3, and R.
- PPT (or PP), polypurine tract is the primer for plus-strand DNA synthesis during reverse transcription.
- U3 is a sequence between PPT and R, which has signal that provirus can use in transcription.
- R is the terminal repeated sequence at 3' end, the same as the R (i.e., repeat region of the 5' end).
Inbetween the 5' and 3' region is the protein encoding region of the retrovirus, consisting of gag proteins, protease (PR), pol proteins and env proteins. Gag proteins are major components of the viral capsid, which are about 2,000-4,000 copies per virion. Protease is expressed differently in different viruses. It functions in proteolytic cleavages during virion maturation to make mature gag and pol proteins. Pol proteins, such as the reverse transcriptase (RT), are responsible for synthesis of viral DNA and integration into host DNA after infection. Finally, env proteins play a role in association and entry of virion into the host cell. Possessing a functional copy of an env gene is what makes retroviruses distinct from retroelements. The env gene serves three distinct functions: enabling the retrovirus to enter/exit host cells through endosomal membrane trafficking, protection from the extracellular environment via the lipid bilayer, and the ability to enter cells. The ability of the retrovirus to bind to its target host cell using specific cell-surface receptors is given by the surface component (SU) of the env, while the ability of the retrovirus to enter the cell via membrane fusion is imparted by the membrane-anchored trans-membrane component (TM). Thus, the env protein is what enables the retrovirus to be infectious.
Please refer to the figure, in it you see all the elements of a retroviral genome and how they interact to contribute to retroviral reverse transcription and integration .
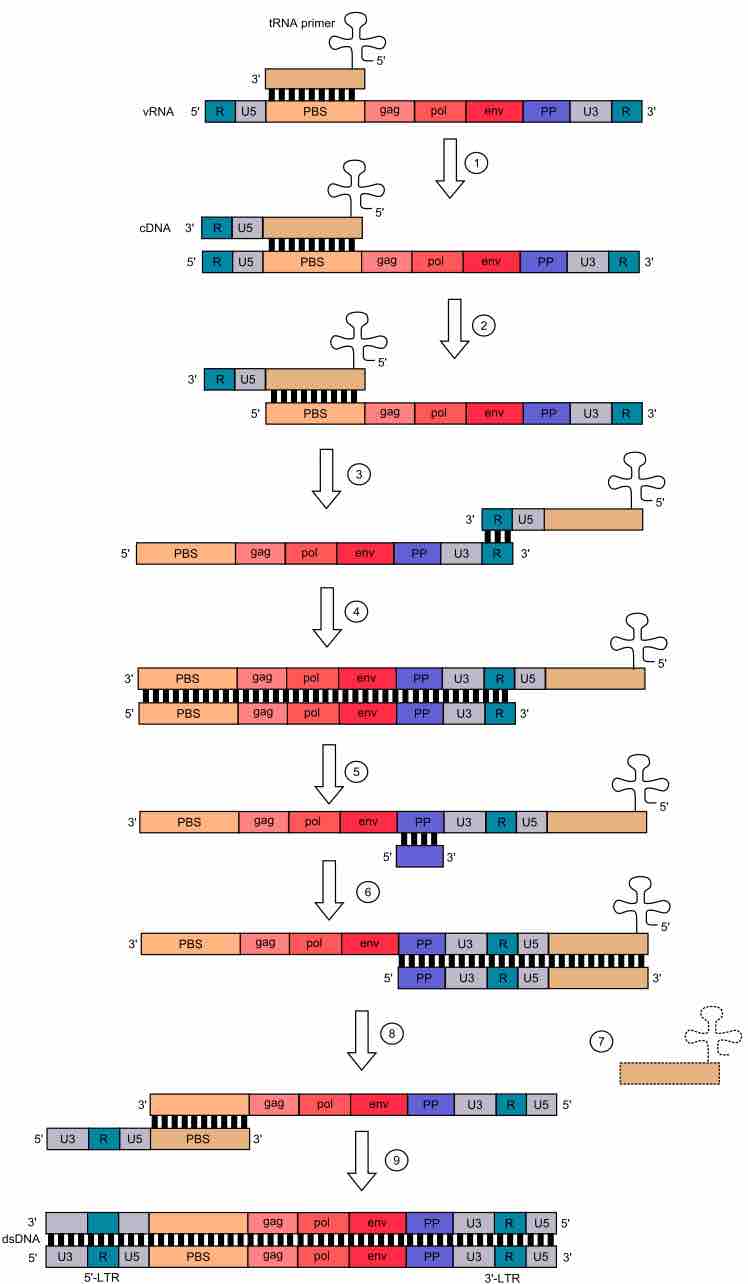
Retroviral genome.
Through the mechanism of reverse transcription by human immunodeficiency virus (HIV), we see what the different genomic elements of a retrovirus are. Reverse transcription occurs in the cytoplasm of host cell. In this process, viral ssRNA is transcribed by the viral reverse transcriptase (RT) into double stranded DNA. Reverse transcription takes place in 5'→3' direction. tRNA ("cloverleaf") hybridizes to PBS and provides -OH group for initiation of reverse transcription. 1) Complementary DNA (cDNA) is formed. 2) Template in RNA:DNA hybrid is degraded by RNase H domain of reverse transcriptase 3) DNA:tRNA is transferred to the 3'-end of the template. 4) First strand synthesis takes place. 5) The rest of viral ssRNA is degraded by RNase H, except for the PP site. 6) Synthesis of second strand of ssDNA is initiated from the 3'-end of the template. tRNA is necessary to synthesis of complementary PBS 7) tRNA is degraded 8) PBS from the second strand hybridizes with the complementary PBS on the first strand. 9) Synthesis of both strands is completed by the DNA Polmerase function of reverse transcriptase. Both dsDNA ends have U3-R-U5 sequences, so called long terminal repeat sequences (3'LTR and 5'LTR, respectively). LTRs mediate integration of the retroviral DNA into another region of the host genome.Key: U3 - promoter region, U5 - recognition site for viral integrase; PBS - primer binding site; PP - polypurine section (polypurine tract); gag, pol, and env. Colors mark complementary sequences.