Describing Diversity
Classification seeks to describe the diversity of bacterial species by naming and grouping organisms based on similarities. Bacteria can be classified on the basis of cell structure, cellular metabolism, or by differences in cell components such as DNA, fatty acids, pigments, antigens, and quinones.
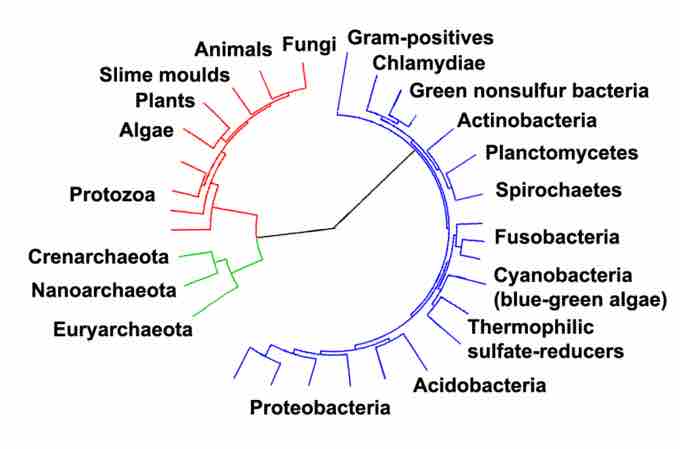
Archaea and other domains
Phylogenetic tree showing the relationship between the Archaea and other domains of life. Eukaryotes are colored red, archaea green and bacteria blue.
While these schemes previously allowed the identification and classification of bacterial strains, it was long unclear whether these differences represented variation between distinct species or between strains of the same species. This uncertainty resulted from the lack of distinctive structures in most bacteria, as well as lateral gene transfer that occurred between unrelated species. Because of the existence of lateral gene transfer, some closely related bacteria have very different morphologies and metabolisms.
To overcome these uncertainties, modern bacterial classification emphasizes molecular systematics, using genetic techniques such as guanine cytosine ratio determination, genome-genome hybridization, as well as sequencing genes that have not undergone extensive lateral gene transfer, such as the rRNA gene.
Uni-Species Cultures for Identification
While there are several molecular tools that allow us to classify or distinguish different bacterial species, this is predicated on obtaining uni-species cultures of a given bacteria. Culture techniques are designed to promote the growth and identify particular bacteria, while restricting the growth of the other bacteria in the sample. Often these techniques are designed for specific specimens; for example, a sputum sample will be treated to identify organisms that cause pneumonia, while stool specimens are cultured on selective media to identify organisms that cause diarrhoea while preventing growth of non-pathogenic bacteria. Specimens that are normally sterile, such as blood, urine, or spinal fluid, are cultured under conditions designed to grow all possible organisms. Once a pathogenic organism has been isolated, it can be further characterized by its morphology, by growth patterns such as aerobic or anaerobic growth, by patterns of hemolysis and by staining. If a bacteria can not be cultured, classification can prove to be very difficult.
DNA Sequencing
However, recent advances in molecular techniques do allow the sequencing of DNA from bacterial species, without the reliance on a pure culture of that given bacteria. Diagnostics using such DNA-based tools, such as polymerase chain reaction, are increasingly popular due to their specificity and speed, compared to culture-based methods. These methods also allow the detection and identification of "viable but nonculturable" cells that are metabolically active but non-dividing, which can be applied to isolates of bacterial species that cannot be cultured. However, even using these improved methods, the total number of bacterial species is not known and cannot even be estimated with any certainty. Following present classification, there are a little less than 9,300 known species of prokaryotes, which includes bacteria and archaea. Attempts to estimate the true level of bacterial diversity have ranged from 107 to 109 total species – and even these diverse estimates may be off by many orders of magnitude.