Virus classification is the process of naming viruses and placing them into a taxonomic system. Much like the classification systems used for cellular organisms, virus classification is the subject of ongoing debate and proposals. This is mainly due to the pseudo-living nature of viruses, which is to say they are non-living particles with some chemical characteristics similar to those of life. As such, they do not fit neatly into the established biological classification system in place for cellular organisms.
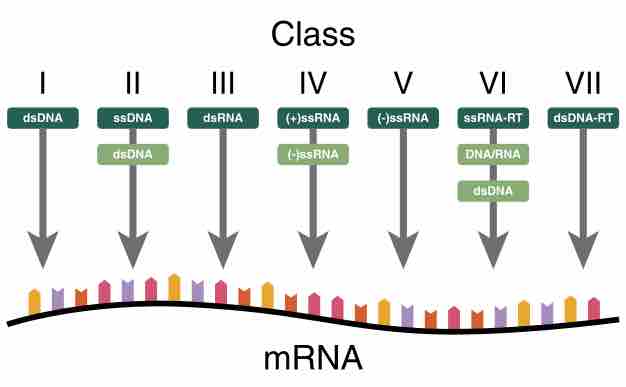
Baltimore classification (first defined in 1971) is a classification system that places viruses into one of seven groups depending on a combination of their nucleic acid (DNA or RNA), strandedness (single-stranded or double-stranded), Sense, and method of replication . Named after David Baltimore, a Nobel Prize-winning biologist, these groups are designated by Roman numerals and discriminate viruses depending on their mode of replication and genome type. Other classifications are determined by the disease caused by the virus or its morphology, neither of which are satisfactory due to different viruses either causing the same disease or looking very similar. In addition, viral structures are often difficult to determine under the microscope.
Viral classes
Baltimore classification of viruses
Classifying viruses according to their genome means that those in a given category will all behave in a similar fashion, offering some indication of how to proceed with further research. Viruses can be placed in one of the seven following groups:
- I: dsDNA viruses (e.g. Adenoviruses, Herpesviruses, Poxviruses)
- II: ssDNA viruses (+)sense DNA (e.g. Parvoviruses)
- III: dsRNA viruses (e.g. Reoviruses)
- IV: (+)ssRNA viruses (+)sense RNA (e.g. Picornaviruses, Togaviruses)
- V: (−)ssRNA viruses (−)sense RNA (e.g. Orthomyxoviruses, Rhabdoviruses)
- VI: ssRNA-RT viruses (+)sense RNA with DNA intermediate in life-cycle (e.g. Retroviruses)
- VII: dsDNA-RT viruses (e.g. Hepadnaviruses)