Virus Classification
To understand the features shared among different groups of viruses, a classification scheme is necessary. However, most viruses are not thought to have evolved from a common ancestor, so the methods that scientists use to classify living things are not very useful. Biologists have used several classification systems in the past, based on the morphology and genetics of the different viruses. However, these earlier classification methods grouped viruses based on which features of the virus they were using to classify them. The most commonly-used classification method today is called the Baltimore classification scheme which is based on how messenger RNA (mRNA) is generated in each particular type of virus. The surface structure of virions can be observed by both scanning and transmission electron microscopy, whereas the internal structures of the virus can only be observed in images from a transmission electron microscope.
Past Systems of Classification
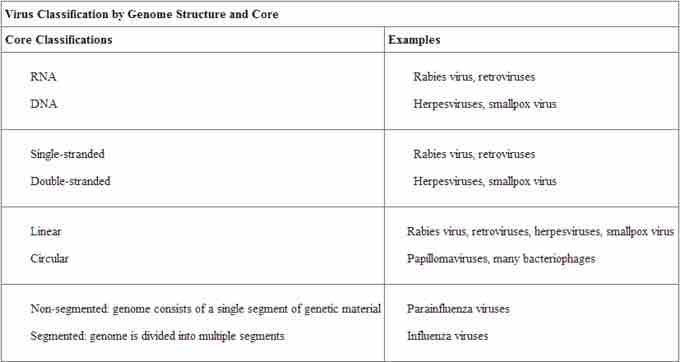
Viruses are classified in several ways: by factors such as their core content, the structure of their capsids, and whether they have an outer envelope. Viruses may use either DNA or RNA as their genetic material. The virus core contains the genome or total genetic content of the virus. Viral genomes tend to be small, containing only those genes that encode proteins that the virus cannot obtain from the host cell. This genetic material may be single- or double-stranded. It may also be linear or circular. While most viruses contain a single nucleic acid, others have genomes that have several, which are called segments. The type of genetic material (DNA or RNA) and its structure (single- or double-stranded, linear or circular, and segmented or non-segmented) are used to classify the virus core structures .
Virus classification by genome structure and core
The type of genetic material (DNA or RNA) and its structure (single- or double-stranded, linear or circular, and segmented or non-segmented) are used to classify the virus core structures.
Viruses can also be classified by the design of their capsids . Isometric viruses have shapes that are roughly spherical, such as poliovirus or herpesviruses. Enveloped viruses have membranes surrounding capsids. Animal viruses, such as HIV, are frequently enveloped. Head and tail viruses infect bacteria and have a head that is similar to icosahedral viruses and a tail shape like filamentous viruses. Capsids are classified as naked icosahedral, enveloped icosahedral, enveloped helical, naked helical, and complex . For example, the tobacco mosaic virus has a naked helical capsid . The adenovirus has an icosahedral capsid .
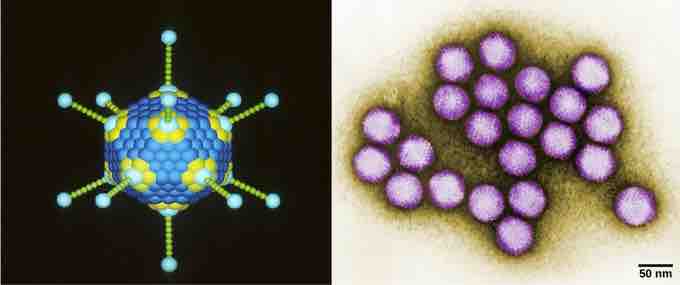
Adenovirus classification
Adenovirus (left) is depicted with a double-stranded DNA genome enclosed in an icosahedral capsid that is 90–100 nm across. The virus, shown clustered in the micrograph (right), is transmitted orally and causes a variety of illnesses in vertebrates, including human eye and respiratory infections.
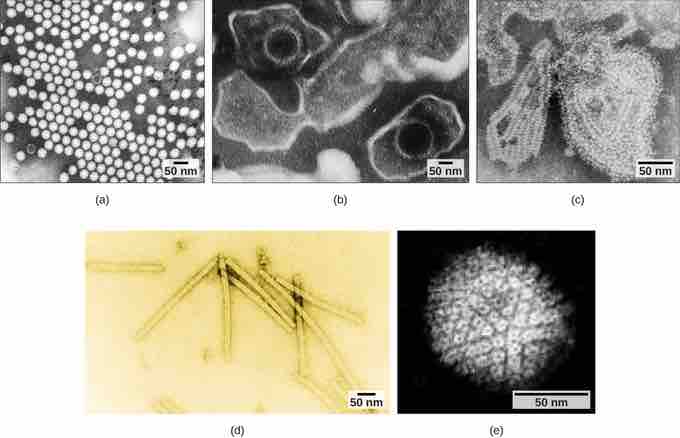
Transmission electron micrograph of viruses
Transmission electron micrographs of various viruses show their structures. The capsid of the (a) polio virus is naked icosahedral; (b) the Epstein-Barr virus capsid is enveloped icosahedral; (c) the mumps virus capsid is an enveloped helix; (d) the tobacco mosaic virus capsid is naked helical; and (e) the herpesvirus capsid is complex.
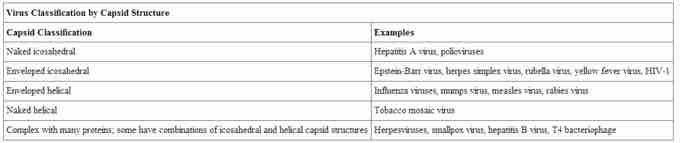
Virus classification by capsid structure
Viruses can also be classified by the design of their capsids which are classified as naked icosahedral, enveloped icosahedral, enveloped helical, naked helical, and complex.
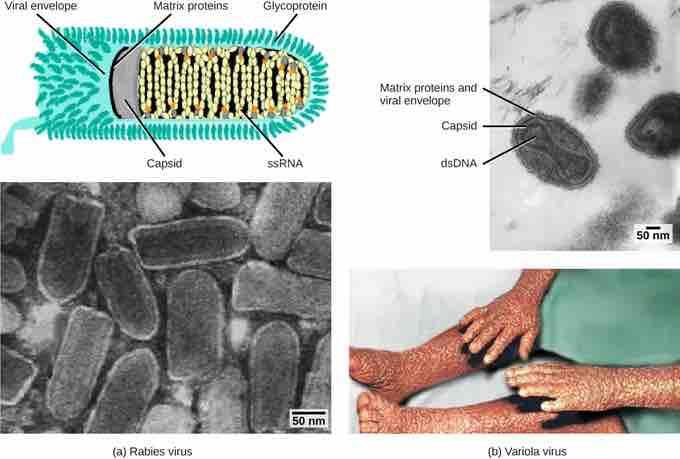
Example of viruses classified by caspid design
Viruses are classified based on their core genetic material and capsid design. (a) Rabies virus has a single-stranded RNA (ssRNA) core and an enveloped helical capsid, whereas (b) variola virus, the causative agent of smallpox, has a double-stranded DNA (dsDNA) core and a complex capsid.
Baltimore Classification
The most commonly-used system of virus classification was developed by Nobel Prize-winning biologist David Baltimore in the early 1970s. In addition to the differences in morphology and genetics mentioned above, the Baltimore classification scheme groups viruses according to how the mRNA is produced during the replicative cycle of the virus. Viruses can contain double-stranded DNA (dsDNA), single-stranded DNA (ssDNA), double-stranded RNA (dsRNA), single-stranded RNA with a positive polarity (ssRNA), ssRNA with a negative polarity, diploid (two copies) ssRNA, and partial dsDNA genomes. Positive polarity means that the genomic RNA can serve directly as mRNA and a negative polarity means that their sequence is complementary to the mRNA .

Baltimore classification
The Baltimore classification scheme, the most commonly used, was developed by Nobel Prize-winning biologist David Baltimore in the early 1970s. The scheme groups viruses according to how the mRNA is produced during the replicative cycle of the virus, in addition to the differences in morphology and genetics.